DLink¶
-
class
pylayers.simul.link.DLink(**kwargs)[source]¶ Bases:
pylayers.simul.link.LinkDeterministic Link Class
- LLayout
Layout to be used
- AaAntenna
Antenna of device dev_a
- AbAntenna
Antenna of device dev_b
- anp.ndarray (3,)
position of a device dev_a
- bnp.ndarray (3,)
position of a device dev_b
- caint
cycle a number
- cbint
cycle b number
- Tanp.ndarray (3,3)
Rotation matrice of Antenna of device dev_a relative to global Layout scene
- Tbnp.ndarray (3,3)
Rotation matrice of Antenna of device dev_b relative to global Layout scene
- fGHznp.ndarray (Nf,)
frequency range of Nf points used for evaluation of channel
- wavWaveform
Waveform to be applied on the channel
- save_idxint
number to identify the h5 file generated
Attributes Summary
Methods Summary
OnClick(event)adp([imax])construct the angular delay profile
afp(**kwargs)Evaluate angular frequency profile
array_exist(key, array[, tol])check an array key has already been stored in h5py file
check_grpname(key, grpname)Check if the key’s data with a given groupname
checkh5()check existence of previous simulations run with the same parameters.
eval(**kwargs)link evaluation
evalH(**kwargs)evaluate channel transfer function
Determine the data group name for the given configuration
get_idx(key, array[, tol])get the index of the requested array in the group key
inforay(iray)provide full information about a specified ray
init_positions([force])initialize random positions for a link
load(obj, grpname, **kwargs)Load a given object in the correct grp
plt_aspread(**kwargs)plot angular spread
plt_cir(**kwargs)plot link channel impulse response
plt_doa(**kwargs)plot direction of arrival and departure
plt_dod(**kwargs)plot direction of arrival and departure
plt_dspread(**kwargs)plot delay spread
reset configuration when a new layout is loaded
save(obj, key, grpname[, force])Save a given object in the correct group
save_init(filename_long)initialize the hdf5 file for link saving
select()show(**kwargs)show the link
stack(key, array)stack new array in h5py file
Attributes Documentation
-
Aa¶
-
Ab¶
-
L¶
-
Lname¶
-
Ta¶
-
Tb¶
-
a¶
-
b¶
-
ca¶
-
cb¶
-
cutoff¶
-
delay_excess_max_ns¶
-
fGHz¶
-
threshold¶
-
wav¶
Methods Documentation
-
afp(**kwargs)[source]¶ Evaluate angular frequency profile
- fGHznp.array
frequency range
az : azimuth angle (radian) tilt : tilt angle (-pi/2<tilt<pi/2) polar : string win : string ‘rect’ | ‘hamming’ _filemeas : string _filecal : string ang_offset : BW : float
bandwidth
- extstring
‘txt’ | ‘mat’
- dirmeasstring
directory of the data in the project path
If a measurement file is given the angular range is obtained from the measurement otherwise the variable az is used.
-
array_exist(key, array, tol=0.001)[source]¶ check an array key has already been stored in h5py file
- key: string
key of the h5py group
- arraynp.ndarray
array type to check existency
- tolnp.float64
tolerance (in meter for key == ‘p_map’)
(ua)
- uanp.ndarray
the indice in the array of the file[key] group if the array is emtpy, value doesn’t exist
Add a tolerance on the rotation angle (T_map)
-
check_grpname(key, grpname)[source]¶ - Check if the key’s data with a given groupname
already exists in the h5py file
- key: string
key of the h5py group
- grpnamestring
groupe name of the h5py file
update the key grpname of self.dexist[key] dictionnary
-
checkh5()[source]¶ check existence of previous simulations run with the same parameters.
update self.dexist dictionnary
-
eval(**kwargs)[source]¶ link evaluation
- applywav :boolean
Apply waveform to H
- forcelist
Force the computation ([‘sig’,’ray2’,’ray,’Ct’,’H’]) AND save (replace previous computations)
- alg1|’old’|’exp’|’exp2’
version of run for signature
- si_progress: bollean ( False)
display progression bar for signatures
- diffractionboolean (False)
takes into consideration diffraction points
- ra_number_mirror_cfint
rays.to3D number of ceil/floor reflexions
- ra_ceil_H: float, (default [])
- ceil height .
If [] : Layout max ceil height If 0 : only floor reflection (outdoor case) If -1 : neither ceil nor floor reflection (2D case)
- ra_vectorized: boolean (True)
if True used the (2015 new) vectorized approach to determine 2drays
- progressbar: str
None: no progress bar python : progress bar in ipython
update self.ak and self.tk
- self.akndarray
alpha_k
- self.tkndarray
tau_k
>>> from pylayers.simul.link import * >>> DL=DLink(L="defstr.lay") >>> DL.eval() >>> DL.show() >>> DL.plt_cir() >>> plt.show()
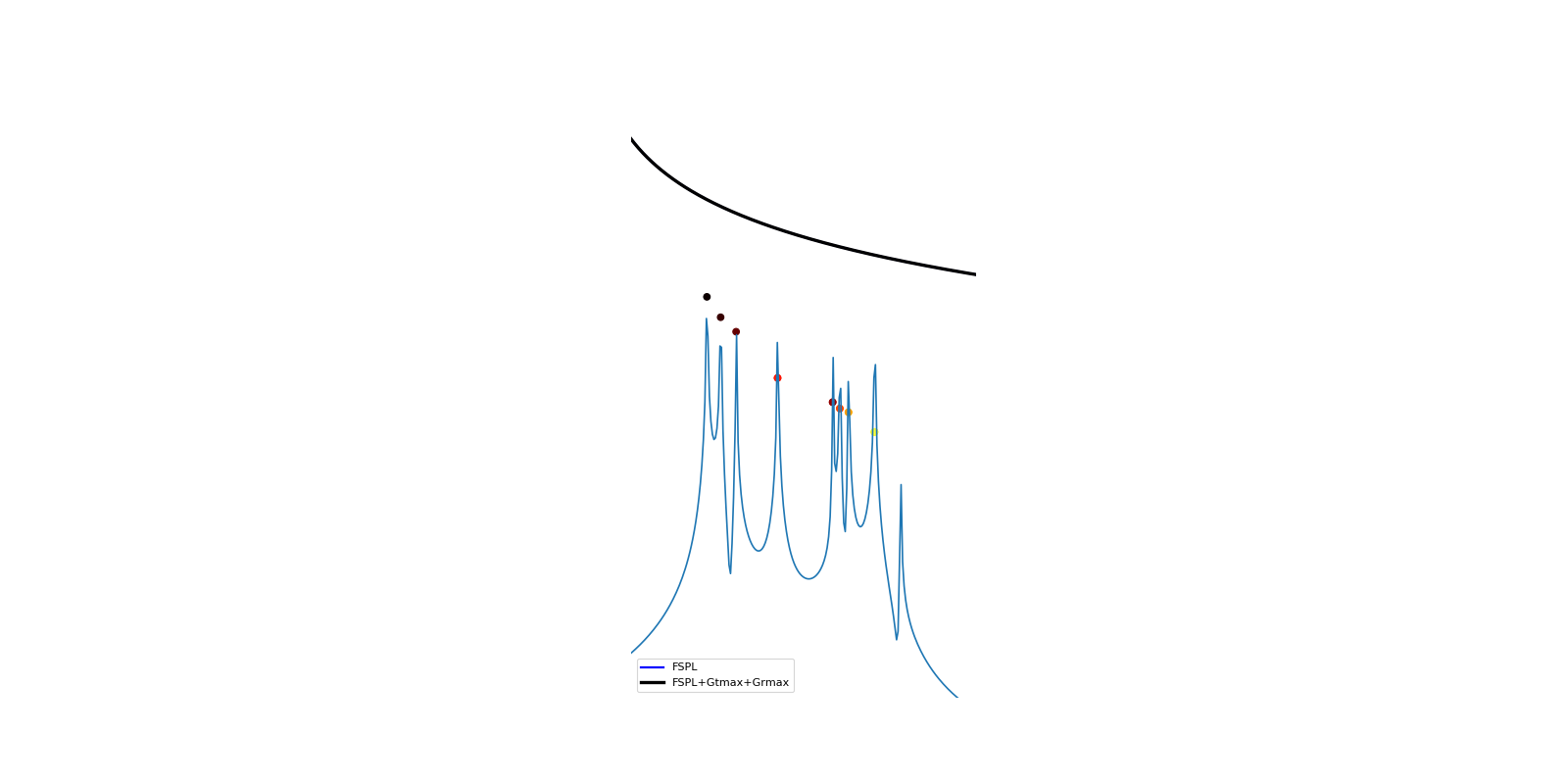
pylayers.antprop.signature pylayers.antprop.rays
-
evalH(**kwargs)[source]¶ evaluate channel transfer function
This function modifies the orientation of the antenna at both sides via Ta and Tb 3x3 matrices and recalculates the channel transfer function for those new orientations. The self.H variable is updated
-
get_grpname()[source]¶ Determine the data group name for the given configuration
Update the key grpname of self.dexist[key] dictionnary, where key = ‘sig’|’ray’|’Ct’|’H’
-
get_idx(key, array, tol=0.001)[source]¶ - get the index of the requested array in the group key
of the hdf5 file. If array doesn’t exist, the hdf5file[key] array is stacked
- key: string
key of the h5py group
- arraynp.ndarray
array to check existence
- tolnp.float64
tolerance (in meters for key == ‘p_map’)
(u_opt, u): tuple
- unp.ndarray
the index in the array of the file[key] group
- u_optstring (‘r’|’s’)
return ‘r’ if array has been read into h5py file return ‘s’ if array has been stacked into the array of group key
Links.array_exist
-
load(obj, grpname, **kwargs)[source]¶ Load a given object in the correct grp
- objObject
(Signatures|Rays|Ctilde|Tchannel)
- grpnamestring
group name of the h5py file
kwargs : layout for sig and rays
-
plt_cir(**kwargs)[source]¶ plot link channel impulse response
BWGHz : Bandwidth Nf : Number of frequency points fftshift : boolean rays : boolean
display rays contributors
- fsplboolean
display free space path loss
fig,ax
pylayers.antprop.channel.Tchannel.getcir
-
plt_doa(**kwargs)[source]¶ plot direction of arrival and departure
fig : plt.figure ax : plt.axis phi: tuple (-180, 180)
phi angle
- normalize: bool
energy normalized
- reversebool
inverse theta and phi represenation
- polarbool
polar representation
cmap: matplotlib.cmap mode: ‘center’ | ‘mean’ | ‘in’
see bsignal.energy
- sfloat
scatter dot size
fontsize: float edgecolors: bool colorbar: bool title : bool
pylayers.antprop.channel.Tchannel.plotd
-
plt_dod(**kwargs)[source]¶ plot direction of arrival and departure
fig : plt.figure ax : plt.axis phi: tuple (-180, 180)
phi angle
- normalize: bool
energy normalized
- reversebool
inverse theta and phi represenation
- polarbool
polar representation
cmap: matplotlib.cmap mode: ‘center’ | ‘mean’ | ‘in’
see bsignal.energy
- sfloat
scatter dot size
fontsize: float edgecolors: bool colorbar: bool title : bool
pylayers.antprop.channel.Tchannel.plotd
-
save(obj, key, grpname, force=False)[source]¶ Save a given object in the correct group
- objObject
(Signatures|Rays|Ctilde|Tchannel)
- keystring
key of the h5py file
- gpnamestring
groupe name of the h5py file
force : boolean or list
-
save_init(filename_long)[source]¶ initialize the hdf5 file for link saving
- filename_longstr
complete path and filename
‘sig’ : Signatures ‘ray2’ : 2D rays ‘ray’ : 3D rays ‘Ct’ : Propagation channel ‘H’ : Transmission channel ‘p_map’ : points ‘c_map’ : cycles ‘f_map’ : frequency ‘A_map’ : antennas ‘T_map’ : rotation
-
show(**kwargs)[source]¶ show the link
- sint
size of Tx/Rx marker in points
- castring
color of termination a (A)
- cbstring
color of termination b (B)
- markerastring
“^”
- markerbstring
“o”
- alphafloat
marker transparency (0 < alpha <1)
- axisboolean
display axis boolean (default True)
- figsizetuple
figure size if fig not specified default (20,10)
- fontsizeint
default 20
- raysboolean
activation of rays vizalization (True)
- bsigboolean
activation of signature vizualization (False)
- bsaveboolean
save in a file indexed by ix
- laddrlist
list of signature addresses
cmap : colormap pol : string
‘tt’,’pp’,’tp’,’pt’,’co’,’cross’,tot’
- labelsboolean
enabling edge label (useful for signature identification)
- colorstring
‘cmap’
linewidth : float alpha : float radius : float
radius in meters for layout vizualization
- dBboolean
default True
- dynfloat
dynamic in dB (def 70dB)
ix vmin vmax bcolorbar : boolean
fig,ax
>>> from pylayers.simul.link import * >>> DL=Link() >>> DL.show(lr=-1,rays=True,dB=True,col='cmap',cmap = plt.cm.jet) >>> DL.show(laddr=[(6,2)],bsig=True)