Contents
- 1 pylayers.util.project Module
- 2 pylayers.gis.layout Module
- 3 pylayers.gis.selectl Module
- 4 pylayers.gis.srtm Module
- 5 pylayers.gis.osmparser Module
- 6 pylayers.gis.ezone Module
- 7 pylayers.antprop.antenna Module
- 8 pylayers.antprop.aarray Module
- 9 pylayers.antprop.spharm Module
- 10 pylayers.antprop.antssh Module
- 11 pylayers.antprop.antvsh Module
- 12 pylayers.antprop.slab Module
- 13 pylayers.antprop.signature Module
- 14 pylayers.antprop.interactions Module
- 15 pylayers.antprop.diffraction Module
- 16 pylayers.antprop.diffRT Module
- 17 pylayers.antprop.rays Module
- 18 pylayers.antprop.loss Module
- 19 pylayers.antprop.channel Module
- 20 pylayers.antprop.loss Module
- 21 pylayers.antprop.coverage Module
- 22 pylayers.antprop.coeffModel Module
- 23 pylayers.simul.link Module
- 24 pylayers.simul.exploit Module
- 25 pylayers.simul.exploit_simulnet Module
- 26 pylayers.simul.simulnet Module
- 27 pylayers.simul.simultraj Module
- 28 pylayers.exploit.simnet Module
- 29 pylayers.measures.mesuwb Module
- 30 pylayers.measures.mesmimo Module
- 31 pylayers.measures.cormoran Module
- 32 pylayers.measures.vna.E5072A Module
- 33 pylayers.measures.parker.smparker Module
- 34 pylayers.signal.bsignal Module
- 35 pylayers.signal.standard Module
- 36 pylayers.signal.device Module
- 37 pylayers.signal.DF Module
- 38 pylayers.signal.waveform Module
- 39 pylayers.mobility.agent Module
- 40 pylayers.mobility.ban.body Module
- 41 pylayers.measures.cormoran Module
1 pylayers.util.project Module¶
1.2 Class Inheritance Diagram¶

2 pylayers.gis.layout Module¶
2.1 Functions¶
PolygonPatch(polygon, **kwargs) |
Constructs a matplotlib patch from a geometric object |
array(object[, dtype, copy, order, subok, ndmin]) |
Create an array. |
cascaded_union |
Returns the union of a sequence of geometries |
cpu_count |
Returns the number of CPUs in the system |
outputGi_func(args) |
|
outputGi_func_test(args) |
|
pbar(verbose, **kwargs) |
|
read_gpickle(path) |
Read graph object in Python pickle format. |
urlopen(url[, data, timeout, cafile, …]) |
|
write_gpickle(G, path[, protocol]) |
Write graph in Python pickle format. |
2.3 Class Inheritance Diagram¶

3 pylayers.gis.selectl Module¶
3.1 Classes¶
RectangleSelector(ax, onselect[, drawtype, …]) |
Select a rectangular region of an axes. |
SelectL(L, fig, ax) |
Associates a Layout and a figure |
3.2 Class Inheritance Diagram¶

4 pylayers.gis.srtm Module¶
4.1 Classes¶
HTMLParser(*[, convert_charrefs]) |
Find tags and other markup and call handler functions. |
InvalidTileError(lat, lon) |
Raised when the SRTM tile file contains invalid data. |
NoSuchTileError(lat, lon) |
Raised when there is no tile for a region. |
SRTMDownloader([server, directory, …]) |
Automatically download SRTM tiles. |
SRTMTile(f, lat, lon) |
Base class for all SRTM tiles. |
WrongTileError(tile_lat, tile_lon, req_lat, …) |
Raised when the value of a pixel outside the tile area is reque sted. |
parseHTMLDirectoryListing() |
4.2 Class Inheritance Diagram¶
5 pylayers.gis.osmparser Module¶
Module OSMParser
5.1 Functions¶
buildingsparse(filename) |
parse buildings |
extract(alat, alon, fileosm, fileout) |
extraction of an osm sub region using osmconvert |
getbdg(fileosm[, verbose]) |
get building from osm file |
getosm(**kwargs) |
get osm region from osmapi |
urlopen(url[, data, timeout, cafile, …]) |
5.2 Classes¶
Basemap([llcrnrlon, llcrnrlat, urcrnrlon, …]) |
|
Coords([idx, latlon]) |
Coords describes a set of points in OSM |
FloorPlan(rootid, coords, nodes, ways, relations) |
FloorPlan class derived from nx.DigGraph |
Nodes |
osm Nodes container |
OsmApi([username, password, passwordfile, …]) |
Main class of osmapi, instanciate this class to use osmapi |
PolyCollection(verts[, sizes, closed]) |
|
PyLayers |
Generic PyLayers Meta Class |
Relations |
|
Way(refs, tags, coords) |
A Way is a polyline or a Polygon (if closed) |
Ways |
Attributes ———- |
5.3 Class Inheritance Diagram¶
6 pylayers.gis.ezone Module¶
6.1 Functions¶
arr2lp(arr) |
convert zeros separated array to list of array |
colorbar(mappable[, cax, ax]) |
Create a colorbar for a ScalarMappable instance. |
conv(extent, m[, mode]) |
convert zone to cartesian or lon lat |
ctrad2qt(extent) |
convert center,radius into a list of qt regions |
dectile([prefix]) |
decode tile name |
distance_on_earth(lat1, long1, lat2, long2) |
Compute great circle distance (the shortest distance over the earths surface) between 2 points on earth: A(lat1,lon1) and B(lat2,lon2) |
dqt(ud16, lL0) |
decode quad tree integer to lon Lat |
enctile(lon, lat) |
encode tile prefix from (lon,lat) |
ent(lL) |
encode lon Lat in natural integer |
eqt(lL) |
encode lon Lat in quad tree integer |
expand(A) |
expand numpy array |
ext2qt([extent, lL0]) |
convert an extent region into a list of qt regions |
get_google_elev_profile(node0, node1[, …]) |
return elevation profile between 2 nodes using google elevation API data |
get_osm_elev_profile(node0, node1[, nb_samples]) |
return elevation profile between 2 nodes using google elevation API data |
haversine(lat1, lon1, lat2, lon2[, mode]) |
lat 1 (Na) lon 1 (Na) lat 2 (Nb) lon 2 (Nb) |
lL2ext(lL) |
convert a lonLat into its qt extent |
lab2ext(lab, lL0) |
label to extent |
make_axes_locatable(axes) |
|
maxloc(f[, threshold]) |
determine local maximum above a threshold |
minsec2dec(old) |
convert latlon from DMS (minute second) to DD (decimal) |
zone(pt[, rm]) |
extract a region from a point and a radius |
6.2 Classes¶
Basemap([llcrnrlon, llcrnrlat, urcrnrlon, …]) |
|
DEM(prefix) |
Class Digital Elevation Model |
Ezone(prefix) |
Earth zone |
Polygon([shell, holes]) |
A two-dimensional figure bounded by a linear ring |
PyLayers |
Generic PyLayers Meta Class |
6.3 Class Inheritance Diagram¶
7 pylayers.antprop.antenna Module¶
7.1 Functions¶
AFLegendre(N, M, x) |
calculate Pmm1n and Pmp1n |
AFLegendre2(L, M, x) |
calculate Pmm1l and Pmp1l |
AFLegendre3(L, M, x) |
calculate Pmm1l and Pmp1l |
BeamGauss(theta, phi[, Gmax, HPBW_az, …]) |
Beam with a Gaussian shape |
CartToSphere(theta, phi, ex, ey, ez[, …]) |
Convert from Cartesian to Spherical |
F0(nu, sigma) |
F0 function for horn antenna pattern |
F1(nu, sigma) |
F1 function for horn antenna pattern |
RepAzimuth1(Ec, theta, phi[, th, typ]) |
response in azimuth |
SSHFunc(L, theta, phi) |
ssh function |
SSHFunc2(L, theta, phi) |
ssh function version 2 |
VW(l, m, theta, phi) |
evaluate vector Spherical Harmonics basis functions |
VW0(n, m, x, phi, Pmm1n, Pmp1n) |
evaluate vector Spherical Harmonics basis functions |
VW2(l, m, x, phi, Pmm1l, Pmp1l) |
evaluate vector Spherical Harmonics basis functions |
cformat(x, y, **kwargs) |
complex format |
compdiag(k, A, th, ph, Fthr, Fphr[, typ, …]) |
makes comparison between original pattern and reconstructed pattern |
cylinder(fig, pa, pb, R) |
plot a cylinder |
displot(pt, ph[, arrow]) |
discontinuous plot |
factorial(*args, **kwds) |
factorial is deprecated! Importing factorial from scipy.misc is deprecated in scipy 1.0.0. |
forcesympol(A) |
plot VSH transform vsh basis in 3D plot |
index_vsh(L, M) |
vector spherical harmonics indexing |
indexssh(L[, mirror]) |
create [l,m] indexation from Lmax |
indexvsh(L) |
calculate index of vsh |
level_energy(A, l[, ifreq, L]) |
calculates energy of the level l |
lmreshape(coeff[, L]) |
level and mode reshaping |
make_axes_locatable(axes) |
|
modeMax(coeff[, L, ifreq]) |
calculates maximal mode |
mode_energy(C, M[, L, ifreq]) |
calculates mode energy |
mode_energy2(A, m[, ifreq, L]) |
calculates mode energy (version 2) |
mulcplot(x, y, **kwargs) |
handling multiple complex variable plots |
plotVW(l, m, theta, phi[, sf]) |
plot VSH transform vsh basis in 3D plot |
pol3D(fig, rho, theta, phi[, sf, shade, title]) |
polar 3D surface plot |
polycol(lpoly[, var]) |
plot a collection of polygon |
rc(group, **kwargs) |
Set the current rc params. |
rectplot(x, xpos[, ylim]) |
plot rectangles on an axis |
relative_error(Eth_original, Eph_original, …) |
calculate relative error between original and model |
shadow(data, ax) |
data : np.array 0 or 1 ax : matplotlib.axes |
show3D(F, theta, phi, k[, col]) |
show 3D matplotlib diagram |
ssh(A[, L, dsf]) |
Parameters ———- |
sshModel(c, d[, L]) |
calculates sshModel |
vsh(A[, dsf]) |
Parameters ———- |
zeros(shape[, dtype, order]) |
Return a new array of given shape and type, filled with zeros. |
7.2 Classes¶
AntPosRot(name, p, T) |
Antenna + position + Rotation |
Antenna([typ]) |
Attributes ———- |
FontProperties([family, style, variant, …]) |
A class for storing and manipulating font properties. |
MaxNLocator(*args, **kwargs) |
Select no more than N intervals at nice locations. |
Pattern() |
Class Pattern |
PolyCollection(verts[, sizes, closed]) |
|
PyLayers |
Generic PyLayers Meta Class |
SCoeff([typ, fmin, fmax, lmax, data, dtype, …]) |
scalar Spherical Harmonics coefficients |
SSHCoeff(Cx, Cy, Cz) |
scalar spherical harmonics |
VCoeff(typ[, fmin, fmax, data, dtype, ind, …]) |
Spherical Harmonics Coefficient |
VSHCoeff(Br, Bi, Cr, Ci) |
Vector Spherical Harmonics Coefficients class |
VectorCoeff(typ[, fmin, fmax, data, dtype, …]) |
class vector spherical harmonics |
7.3 Class Inheritance Diagram¶

8 pylayers.antprop.aarray Module¶
8.1 Functions¶
k2xyz(ik, sh) |
Parameters ———- |
weights(nx, nz, kx, kz, Kx, Kz) |
Practical Demonstration of Limited Feedback Beamforming for mmWave Systems |
xyztok(iz, iy, ix[, Nx, Ny]) |
8.2 Classes¶
AntArray(**kwargs) |
Class AntArray |
Array(p[, w]) |
Array class |
Combiner(Wbr, Whb, Wsh) |
|
Precoder(Fhs, Fbh, Fht) |
|
TXRU() |
Tranceiver Units |
UCArray(p[, w]) |
Uniform Circular Array |
ULArray(**kwargs) |
Uniform Linear Array |
8.3 Class Inheritance Diagram¶
9 pylayers.antprop.spharm Module¶
9.1 Functions¶
AFLegendre(N, M, x) |
calculate Pmm1n and Pmp1n |
AFLegendre2(L, M, x) |
calculate Pmm1l and Pmp1l |
AFLegendre3(L, M, x) |
calculate Pmm1l and Pmp1l |
VW(l, m, theta, phi) |
evaluate vector Spherical Harmonics basis functions |
VW0(n, m, x, phi, Pmm1n, Pmp1n) |
evaluate vector Spherical Harmonics basis functions |
VW2(l, m, x, phi, Pmm1l, Pmp1l) |
evaluate vector Spherical Harmonics basis functions |
cformat(x, y, **kwargs) |
complex format |
cylinder(fig, pa, pb, R) |
plot a cylinder |
displot(pt, ph[, arrow]) |
discontinuous plot |
factorial(*args, **kwds) |
factorial is deprecated! Importing factorial from scipy.misc is deprecated in scipy 1.0.0. |
index_vsh(L, M) |
vector spherical harmonics indexing |
indexssh(L[, mirror]) |
create [l,m] indexation from Lmax |
indexvsh(L) |
calculate index of vsh |
mulcplot(x, y, **kwargs) |
handling multiple complex variable plots |
plotVW(l, m, theta, phi[, sf]) |
plot VSH transform vsh basis in 3D plot |
pol3D(fig, rho, theta, phi[, sf, shade, title]) |
polar 3D surface plot |
polycol(lpoly[, var]) |
plot a collection of polygon |
rc(group, **kwargs) |
Set the current rc params. |
rectplot(x, xpos[, ylim]) |
plot rectangles on an axis |
shadow(data, ax) |
data : np.array 0 or 1 ax : matplotlib.axes |
9.2 Classes¶
FontProperties([family, style, variant, …]) |
A class for storing and manipulating font properties. |
PolyCollection(verts[, sizes, closed]) |
|
PyLayers |
Generic PyLayers Meta Class |
SCoeff([typ, fmin, fmax, lmax, data, dtype, …]) |
scalar Spherical Harmonics coefficients |
SSHCoeff(Cx, Cy, Cz) |
scalar spherical harmonics |
VCoeff(typ[, fmin, fmax, data, dtype, ind, …]) |
Spherical Harmonics Coefficient |
VSHCoeff(Br, Bi, Cr, Ci) |
Vector Spherical Harmonics Coefficients class |
VectorCoeff(typ[, fmin, fmax, data, dtype, …]) |
class vector spherical harmonics |
9.3 Class Inheritance Diagram¶
10 pylayers.antprop.antssh Module¶
10.1 Functions¶
AFLegendre(N, M, x) |
calculate Pmm1n and Pmp1n |
AFLegendre2(L, M, x) |
calculate Pmm1l and Pmp1l |
AFLegendre3(L, M, x) |
calculate Pmm1l and Pmp1l |
CartToSphere(theta, phi, ex, ey, ez[, …]) |
Convert from Cartesian to Spherical |
SSHFunc(L, theta, phi) |
ssh function |
SSHFunc2(L, theta, phi) |
ssh function version 2 |
SphereToCart(theta, phi, eth, eph, bfreq) |
Spherical to Cartesian |
VW(l, m, theta, phi) |
evaluate vector Spherical Harmonics basis functions |
VW0(n, m, x, phi, Pmm1n, Pmp1n) |
evaluate vector Spherical Harmonics basis functions |
VW2(l, m, x, phi, Pmm1l, Pmp1l) |
evaluate vector Spherical Harmonics basis functions |
cformat(x, y, **kwargs) |
complex format |
cylinder(fig, pa, pb, R) |
plot a cylinder |
displot(pt, ph[, arrow]) |
discontinuous plot |
factorial(*args, **kwds) |
factorial is deprecated! Importing factorial from scipy.misc is deprecated in scipy 1.0.0. |
index_vsh(L, M) |
vector spherical harmonics indexing |
indexssh(L[, mirror]) |
create [l,m] indexation from Lmax |
indexvsh(L) |
calculate index of vsh |
mulcplot(x, y, **kwargs) |
handling multiple complex variable plots |
plotVW(l, m, theta, phi[, sf]) |
plot VSH transform vsh basis in 3D plot |
pol3D(fig, rho, theta, phi[, sf, shade, title]) |
polar 3D surface plot |
polycol(lpoly[, var]) |
plot a collection of polygon |
rc(group, **kwargs) |
Set the current rc params. |
rectplot(x, xpos[, ylim]) |
plot rectangles on an axis |
shadow(data, ax) |
data : np.array 0 or 1 ax : matplotlib.axes |
ssh(A[, L, dsf]) |
Parameters ———- |
sshs(G, fGHz, th, ph[, L]) |
scalar spherical harmonics transform |
11 pylayers.antprop.antvsh Module¶
11.1 Functions¶
AFLegendre(N, M, x) |
calculate Pmm1n and Pmp1n |
AFLegendre2(L, M, x) |
calculate Pmm1l and Pmp1l |
AFLegendre3(L, M, x) |
calculate Pmm1l and Pmp1l |
VW(l, m, theta, phi) |
evaluate vector Spherical Harmonics basis functions |
VW0(n, m, x, phi, Pmm1n, Pmp1n) |
evaluate vector Spherical Harmonics basis functions |
VW2(l, m, x, phi, Pmm1l, Pmp1l) |
evaluate vector Spherical Harmonics basis functions |
cformat(x, y, **kwargs) |
complex format |
cylinder(fig, pa, pb, R) |
plot a cylinder |
displot(pt, ph[, arrow]) |
discontinuous plot |
factorial(*args, **kwds) |
factorial is deprecated! Importing factorial from scipy.misc is deprecated in scipy 1.0.0. |
index_vsh(L, M) |
vector spherical harmonics indexing |
indexssh(L[, mirror]) |
create [l,m] indexation from Lmax |
indexvsh(L) |
calculate index of vsh |
mulcplot(x, y, **kwargs) |
handling multiple complex variable plots |
plotVW(l, m, theta, phi[, sf]) |
plot VSH transform vsh basis in 3D plot |
pol3D(fig, rho, theta, phi[, sf, shade, title]) |
polar 3D surface plot |
polycol(lpoly[, var]) |
plot a collection of polygon |
rc(group, **kwargs) |
Set the current rc params. |
rectplot(x, xpos[, ylim]) |
plot rectangles on an axis |
shadow(data, ax) |
data : np.array 0 or 1 ax : matplotlib.axes |
vsh(A[, dsf]) |
Parameters ———- |
12 pylayers.antprop.slab Module¶
12.1 Functions¶
calRT_3layers_model(x, epsr, d, fGHz, theta) |
calculate R and T for an homogeneous Slab |
calRT_homogeneous_model(x, epsr, d, fGHz, theta) |
calculate R and T for an homogeneous Slab |
calsig(cval, fGHz[, typ]) |
evaluate sigma from epsr or index at a given frequency |
12.2 Classes¶
Interface([fGHz, theta, name]) |
Interface between 2 medium |
Mat(name, **dm) |
Handle constitutive materials dictionnary |
MatDB([_fileini, dm]) |
MatDB Class : Material database |
MatInterface(lmat, l, fGHz, theta) |
MatInterface : Class for Interface between two materials |
PyLayers |
Generic PyLayers Meta Class |
Slab(name, matDB[, ds]) |
Handle a Slab |
SlabDB([fileslab, filemat, ds, dm]) |
Slab data base |
interp1d(x, y[, kind, axis, copy, …]) |
Interpolate a 1-D function. |
12.3 Class Inheritance Diagram¶
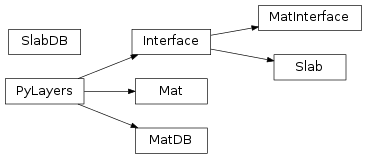
13 pylayers.antprop.signature Module¶
13.1 Functions¶
gidl(g) |
gi without diffraction |
plot_lines(ax, ob[, color]) |
plot lines with colors |
plot_poly(ax, ob[, color]) |
plot polygon |
shLtmp(L) |
|
showsig(L, s[, tx, rx]) |
show signature |
showsig2(lsig, L, tahe) |
|
valid(lsig, L[, tahe]) |
Check if a signature is valid. |
13.2 Classes¶
Axes3D(fig[, rect]) |
3D axes object. |
PyLayers |
Generic PyLayers Meta Class |
Rays(pTx, pRx) |
Class handling a set of rays |
Signature(sig) |
class Signature |
Signatures(L, source, target[, cutoff, …]) |
set of Signature given 2 Gt cycle (convex) indices |
tqdm([iterable, desc, total, leave, file, …]) |
Decorate an iterable object, returning an iterator which acts exactly like the original iterable, but prints a dynamically updating progressbar every time a value is requested. |
13.3 Class Inheritance Diagram¶
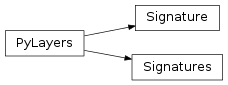
14 pylayers.antprop.interactions Module¶
14.1 Classes¶
IntB([data, dtype, idx, slab]) |
Local Basis interaction class |
IntD([data, dtype, idx, fGHz, slab]) |
diffraction interaction class |
IntR([data, dtype, idx, slab]) |
Reflexion interaction class |
IntT([data, dtype, idx, slab]) |
Transmission interaction class |
Inter([typ, data, dtype, idx, _filemat, …]) |
Interactions |
Interactions([slab]) |
Interaction parameters |
Interface([fGHz, theta, name]) |
Interface between 2 medium |
Mat(name, **dm) |
Handle constitutive materials dictionnary |
MatDB([_fileini, dm]) |
MatDB Class : Material database |
MatInterface(lmat, l, fGHz, theta) |
MatInterface : Class for Interface between two materials |
PyLayers |
Generic PyLayers Meta Class |
Slab(name, matDB[, ds]) |
Handle a Slab |
SlabDB([fileslab, filemat, ds, dm]) |
Slab data base |
interp1d(x, y[, kind, axis, copy, …]) |
Interpolate a 1-D function. |
14.2 Class Inheritance Diagram¶
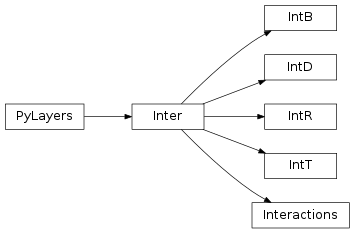
16 pylayers.antprop.diffRT Module¶
16.1 Functions¶
Dfunc(sign, k, N, dphi, si, sd[, xF, F, beta]) |
Parameters ———- |
FreF(x) |
F function from Pathack |
FreF2(x) |
F function using numpy fresnel function |
FresnelI(x) |
calculates Fresnel integral |
G(N, phi0, Ro, Rn) |
grazing angle correction |
R(th, k, er, err, sigma, ur, urr, deltah) |
R coeff |
diff(fGHz, phi0, phi, si, sd, N, mat0, matN) |
Luebbers Diffration coefficient for Ray tracing |
17 pylayers.antprop.rays Module¶
17.1 Classes¶
Ctilde() |
container for the 4 components of the polarimetric ray channel |
IntB([data, dtype, idx, slab]) |
Local Basis interaction class |
IntD([data, dtype, idx, fGHz, slab]) |
diffraction interaction class |
IntR([data, dtype, idx, slab]) |
Reflexion interaction class |
IntT([data, dtype, idx, slab]) |
Transmission interaction class |
Inter([typ, data, dtype, idx, _filemat, …]) |
Interactions |
Interactions([slab]) |
Interaction parameters |
Interface([fGHz, theta, name]) |
Interface between 2 medium |
Layout([arg]) |
Handling Layout |
Mat(name, **dm) |
Handle constitutive materials dictionnary |
MatDB([_fileini, dm]) |
MatDB Class : Material database |
MatInterface(lmat, l, fGHz, theta) |
MatInterface : Class for Interface between two materials |
PyLayers |
Generic PyLayers Meta Class |
Rays(pTx, pRx) |
Class handling a set of rays |
Slab(name, matDB[, ds]) |
Handle a Slab |
SlabDB([fileslab, filemat, ds, dm]) |
Slab data base |
interp1d(x, y[, kind, axis, copy, …]) |
Interpolate a 1-D function. |
17.2 Class Inheritance Diagram¶

18 pylayers.antprop.loss Module¶
18.1 Functions¶
Dgrid_points(points, Px) |
distance point to grid |
Dgrid_zone(zone, Px) |
Distance point to zone |
FMetisShad(fGHz, r, D[, sign]) |
F Metis shadowing function |
FMetisShad2(fGHz, r, D[, sign]) |
F Metis shadowing function |
Loss0(S, rx, ry, f, p) |
calculate Loss through Layers for theta=0 deg |
LossMetisShadowing(fGHz, tx, rx, pg, uw, uh, …) |
Calculate the Loss from |
LossMetisShadowing2(fGHz, tx, rx, pg, uw, …) |
Calculate the Loss from |
Loss_diff(u) |
calculate Path Loss of the diffraction |
Losst(L, fGHz, p1, p2[, dB, bceilfloor]) |
calculate Losses between links p1-p2 |
OneSlopeMdl(D, n, fGHz) |
one slope model |
PL(fGHz, pts, p[, n, dB, d0]) |
calculate Free Space Path Loss |
PL0(fGHz[, GtdB, GrdB, R]) |
Path Loss at frequency fGHZ @ R |
bullington(d, height, fGHz) |
edges attenuation with Bullington method |
calnu(h, d1, d2, fGHz) |
Calculate the diffraction Fresnel parameter |
cdf(x[, colsym, lab, lw]) |
plot the cumulative density function |
cost2100(pMS, pBS, fGHz[, nfloor, dB]) |
cost 2100 model |
cost231(pBS, pMS, hroof, phir, wr, fMHz[, …]) |
Walfish Ikegami model (COST 231) |
cost259(pMS, pBS, fMHz) |
cost259 model |
cover(X, Y, Z, Ha, Hb, fGHz, K[, method]) |
outdoor coverage on a region |
deygout(d, height, fGHz, L, depth) |
Deygout attenuation |
gaspl(d, fGHz, T, PhPa, wvden) |
attenuation due to atmospheric gases |
hata(pMS, pBS, fGHz, hMS, hBS, typ) |
Hata Path loss model |
jit([signature_or_function, locals, target, …]) |
This decorator is used to compile a Python function into native code. |
lossref_compute(P, h0, h1[, k]) |
compute loss and reflection rays on curved earth |
route(X, Y, Z, Ha, Hb, fGHz, K[, method]) |
diffraction loss along a route |
two_rays_curvedearthold(P, h0, h1[, fGHz]) |
Parameters ———- |
two_rays_flatearth(fGHz, **kwargs) |
Parameters ———- |
visuPts(S, nu, nd, Pts, Values[, fig, sp, …]) |
visuPt : Visualization of values a given points |
19 pylayers.antprop.channel Module¶
19.1 Classes¶
ADPchannel([x, dtype, y, dtype, az, dtype, …]) |
Angular Delay Profile channel |
AFPchannel([x, dtype, y, dtype, tx, dtype, …]) |
Angular Frequency Profile channel |
Axes3D(fig[, rect]) |
3D axes object. |
Ctilde() |
container for the 4 components of the polarimetric ray channel |
Mchannel(x, y, **kwargs) |
Handle the measured channel |
PyLayers |
Generic PyLayers Meta Class |
TBchannel([x, dtype, y, dtype, label]) |
radio channel in non uniform delay domain |
TUDchannel([x, dtype, y, dtype, taud, …]) |
Uniform channel in Time domain with delay |
TUchannel([x, dtype, y, dtype, label]) |
Uniform channel in delay domain |
Tchannel([x, y, tau, shape, dtype, dod, …]) |
Handle the transmission channel |
19.2 Class Inheritance Diagram¶
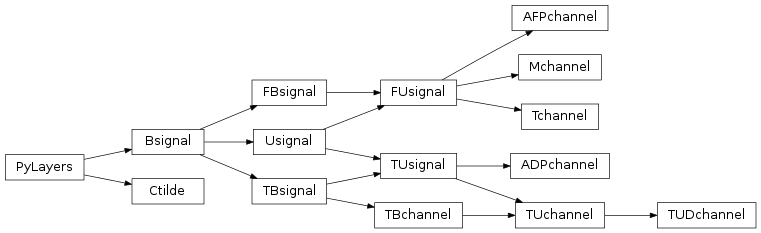
20 pylayers.antprop.loss Module¶
20.1 Functions¶
Dgrid_points(points, Px) |
distance point to grid |
Dgrid_zone(zone, Px) |
Distance point to zone |
FMetisShad(fGHz, r, D[, sign]) |
F Metis shadowing function |
FMetisShad2(fGHz, r, D[, sign]) |
F Metis shadowing function |
Loss0(S, rx, ry, f, p) |
calculate Loss through Layers for theta=0 deg |
LossMetisShadowing(fGHz, tx, rx, pg, uw, uh, …) |
Calculate the Loss from |
LossMetisShadowing2(fGHz, tx, rx, pg, uw, …) |
Calculate the Loss from |
Loss_diff(u) |
calculate Path Loss of the diffraction |
Losst(L, fGHz, p1, p2[, dB, bceilfloor]) |
calculate Losses between links p1-p2 |
OneSlopeMdl(D, n, fGHz) |
one slope model |
PL(fGHz, pts, p[, n, dB, d0]) |
calculate Free Space Path Loss |
PL0(fGHz[, GtdB, GrdB, R]) |
Path Loss at frequency fGHZ @ R |
bullington(d, height, fGHz) |
edges attenuation with Bullington method |
calnu(h, d1, d2, fGHz) |
Calculate the diffraction Fresnel parameter |
cdf(x[, colsym, lab, lw]) |
plot the cumulative density function |
cost2100(pMS, pBS, fGHz[, nfloor, dB]) |
cost 2100 model |
cost231(pBS, pMS, hroof, phir, wr, fMHz[, …]) |
Walfish Ikegami model (COST 231) |
cost259(pMS, pBS, fMHz) |
cost259 model |
cover(X, Y, Z, Ha, Hb, fGHz, K[, method]) |
outdoor coverage on a region |
deygout(d, height, fGHz, L, depth) |
Deygout attenuation |
gaspl(d, fGHz, T, PhPa, wvden) |
attenuation due to atmospheric gases |
hata(pMS, pBS, fGHz, hMS, hBS, typ) |
Hata Path loss model |
jit([signature_or_function, locals, target, …]) |
This decorator is used to compile a Python function into native code. |
lossref_compute(P, h0, h1[, k]) |
compute loss and reflection rays on curved earth |
route(X, Y, Z, Ha, Hb, fGHz, K[, method]) |
diffraction loss along a route |
two_rays_curvedearthold(P, h0, h1[, fGHz]) |
Parameters ———- |
two_rays_flatearth(fGHz, **kwargs) |
Parameters ———- |
visuPts(S, nu, nd, Pts, Values[, fig, sp, …]) |
visuPt : Visualization of values a given points |
21 pylayers.antprop.coverage Module¶
21.1 Classes¶
CheckButtons(ax, labels, actives) |
A GUI neutral radio button. |
Coverage([_fileini]) |
Handle Layout Coverage |
Layout([arg]) |
Handling Layout |
PyLayers |
Generic PyLayers Meta Class |
RadioNode([name, typ, _fileini, _fileant]) |
container for a Radio Node |
Slider(ax, label, valmin, valmax[, valinit, …]) |
A slider representing a floating point range. |
Trajectories() |
Define a list of trajectory |
Trajectory([df, ID, name, typ]) |
define a trajectory |
datetime(year, month, day[, hour[, minute[, …) |
The year, month and day arguments are required. |
product |
product(*iterables, repeat=1) –> product object |
21.2 Class Inheritance Diagram¶

22 pylayers.antprop.coeffModel Module¶
22.1 Functions¶
RepAzimuth1(Ec, theta, phi[, th, typ]) |
response in azimuth |
level_energy(A, l[, ifreq, L]) |
calculates energy of the level l |
lmreshape(coeff[, L]) |
level and mode reshaping |
modeMax(coeff[, L, ifreq]) |
calculates maximal mode |
mode_energy(C, M[, L, ifreq]) |
calculates mode energy |
mode_energy2(A, m[, ifreq, L]) |
calculates mode energy (version 2) |
relative_error(Eth_original, Eph_original, …) |
calculate relative error between original and model |
sshModel(c, d[, L]) |
calculates sshModel |
zeros(shape[, dtype, order]) |
Return a new array of given shape and type, filled with zeros. |
23 pylayers.simul.link Module¶
23.1 Classes¶
AFPchannel([x, dtype, y, dtype, tx, dtype, …]) |
Angular Frequency Profile channel |
Antenna([typ]) |
Attributes ———- |
Ctilde() |
container for the 4 components of the polarimetric ray channel |
DLink(**kwargs) |
Deterministic Link Class |
Layout([arg]) |
Handling Layout |
Link() |
Link class |
PyLayers |
Generic PyLayers Meta Class |
RadioNode([name, typ, _fileini, _fileant]) |
container for a Radio Node |
Rays(pTx, pRx) |
Class handling a set of rays |
SLink() |
|
Signature(sig) |
class Signature |
Signatures(L, source, target[, cutoff, …]) |
set of Signature given 2 Gt cycle (convex) indices |
Tchannel([x, y, tau, shape, dtype, dod, …]) |
Handle the transmission channel |
VTKDataSource |
This source manages a VTK dataset given to it. |
23.2 Class Inheritance Diagram¶
24 pylayers.simul.exploit Module¶
24.1 Classes¶
Axes3D(fig[, rect]) |
3D axes object. |
Bsignal([x, dtype, y, dtype, label]) |
Signal with an embedded time base |
Error |
|
Exploit([simnetfile]) |
class Exploit |
FBsignal([x, dtype, y, dtype, label]) |
FBsignal : Base signal in Frequency domain |
FHsignal([x, dtype, y, dtype, label]) |
FHsignal : Hermitian uniform signal in Frequency domain |
FUsignal([x, dtype, y, dtype, label]) |
FUsignal : Uniform signal in Frequency Domain |
Noise([ti, tf, fsGHz, PSDdBmpHz, NF, R, seed]) |
Create noise |
PolyCollection(verts[, sizes, closed]) |
|
PyLayers |
Generic PyLayers Meta Class |
TBsignal([x, dtype, y, dtype, label]) |
Based signal in Time domain |
TUsignal([x, dtype, y, dtype, label]) |
Uniform signal in time domain |
Usignal([x, dtype, y, dtype, label]) |
Signal with an embedded uniform Base |
24.2 Class Inheritance Diagram¶

25 pylayers.simul.exploit_simulnet Module¶
25.2 Class Inheritance Diagram¶

26 pylayers.simul.simulnet Module¶
26.1 Classes¶
Agent(**args) |
Class Agent |
CheckButtons(ax, labels, actives) |
A GUI neutral radio button. |
EMSolver([L]) |
Invoque an electromagnetic solver |
Gcom(net, sim) |
Communication graph |
Layout([arg]) |
Handling Layout |
Network([owner, EMS, PN]) |
Network class |
Node(**kwargs) |
Class Node |
PNetwork(**args) |
Process version of the Network class |
Process(env, generator) |
Process an event yielding generator. |
PyLayers |
Generic PyLayers Meta Class |
Save(**args) |
Save all variables of a simulnet simulation. |
ShowNet(**args) |
Show network process |
ShowTable(**args) |
Show table process |
Simul() |
Attributes ———- |
Slab(name, matDB[, ds]) |
Handle a Slab |
Slider(ax, label, valmin, valmax[, valinit, …]) |
A slider representing a floating point range. |
Trajectories() |
Define a list of trajectory |
Trajectory([df, ID, name, typ]) |
define a trajectory |
datetime(year, month, day[, hour[, minute[, …) |
The year, month and day arguments are required. |
26.2 Class Inheritance Diagram¶

27 pylayers.simul.simultraj Module¶
27.1 Classes¶
AFPchannel([x, dtype, y, dtype, tx, dtype, …]) |
Angular Frequency Profile channel |
Antenna([typ]) |
Attributes ———- |
Axes3D(fig[, rect]) |
3D axes object. |
Basemap([llcrnrlon, llcrnrlat, urcrnrlon, …]) |
|
Body([_filebody, _filemocap, _filewear, …]) |
Class to manage a Body model |
Button(ax, label[, image, color, hovercolor]) |
A GUI neutral button. |
CheckButtons(ax, labels, actives) |
A GUI neutral radio button. |
CorSer([serie, day, source, layout]) |
Handle CORMORAN measurement data |
Ctilde() |
container for the 4 components of the polarimetric ray channel |
Cursor(ax[, horizOn, vertOn, useblit]) |
A horizontal and vertical line that spans the axes and moves with the pointer. |
DF([b, a]) |
Digital Filter Class |
DLink(**kwargs) |
Deterministic Link Class |
Device([devname, ID, owner, typ]) |
Device Class |
Layout([arg]) |
Handling Layout |
Link() |
Link class |
Network([owner, EMS, PN]) |
Network class |
Pool |
alias of pathos.multiprocessing.ProcessPool |
PyLayers |
Generic PyLayers Meta Class |
RadioNode([name, typ, _fileini, _fileant]) |
container for a Radio Node |
Rays(pTx, pRx) |
Class handling a set of rays |
SLink() |
|
SelectL(L, fig, ax) |
Associates a Layout and a figure |
Signature(sig) |
class Signature |
Signatures(L, source, target[, cutoff, …]) |
set of Signature given 2 Gt cycle (convex) indices |
Simul([source, verbose]) |
Link oriented simulation |
Slider(ax, label, valmin, valmax[, valinit, …]) |
A slider representing a floating point range. |
Tchannel([x, y, tau, shape, dtype, dod, …]) |
Handle the transmission channel |
VTKDataSource |
This source manages a VTK dataset given to it. |
combinations |
combinations(iterable, r) –> combinations object |
partial |
partial(func, *args, **keywords) - new function with partial application of the given arguments and keywords. |
product |
product(*iterables, repeat=1) –> product object |
27.2 Class Inheritance Diagram¶

28 pylayers.exploit.simnet Module¶
28.1 Classes¶
Simnet2cir([simnetfile]) |
This class allows to perform a raytracing simulation from a simulnet simulation. |
Simnet2loc([savefile]) |
Summary ——- |
28.2 Class Inheritance Diagram¶

29 pylayers.measures.mesuwb Module¶
29.1 RAW_DATA Class¶
RAW_DATA.__init__(d) |
Initialize self. |
29.2 CAL_DATA Class¶
CAL_DATA.__init__(d) |
depending of the version of scipy io.loadma do not give the same output |
CAL_DATA.plot() |
|
CAL_DATA.getwave() |
29.4 Tdd Class¶
Tdd.__init__(d) |
Initialize self. |
Tdd.PL(fmin, fmax, B) |
Calculate NB Path Loss on a given UWB frequency band |
Tdd.show([fig, delay, display, title, col, …]) |
show the 4 Impulse Radio Impulse responses |
Tdd.show_span([delay, wide]) |
show span |
Tdd.box() |
evaluate min and max of the 4 channels |
Tdd.plot([type]) |
type : raw | filter |
29.5 TFP Class¶
TFP.__init__() |
Initialize self. |
TFP.append(FP) |
append data to fingerprint |
29.6 FP Class¶
FP.__init__(M, k[, alpha, Tint, sym, nint, …]) |
object constructor |
29.7 UWBMeasure¶
UWBMeasure.__init__([nTx, h, display]) |
object constructor |
UWBMeasure.info() |
|
UWBMeasure.show([fig, delay, display, col, …]) |
show measurement in time domain |
UWBMeasure.Epercent() |
|
UWBMeasure.toa_max2() |
calculate toa_max (meth2) |
UWBMeasure.tau_Emax() |
calculate the delay of energy peak |
UWBMeasure.tau_moy([display]) |
calculate mean excess delay |
UWBMeasure.tau_rms([display]) |
calculate the rms delay spread |
UWBMeasure.toa_new([display]) |
descendant threshold based toa estimation |
UWBMeasure.toa_win([n, display]) |
descendant threshold based toa estimation |
UWBMeasure.toa_max([n, display]) |
descendant threshold based toa estimation |
UWBMeasure.toa_th(r, k[, display]) |
threshold based toa estimation using energy peak |
UWBMeasure.toa_cum(n[, display]) |
threshold based toa estimation using cumulative energy |
UWBMeasure.taumax() |
|
UWBMeasure.Emax([Tint, sym, dB]) |
calculate maximum energy |
UWBMeasure.Etot([toffns, tdns, dB]) |
Calculate total energy for the 4 channels |
UWBMeasure.Efirst([Tint, sym, dB]) |
calculate energy in first path |
UWBMeasure.Etau0([Tint, sym, dB]) |
calculate the energy around delay tau0 |
UWBMeasure.ecdf([Tnoise, rem_noise, …]) |
calculate energy cumulative density function |
UWBMeasure.tdelay() |
build an array with delay values |
UWBMeasure.fp([alpha]) |
build fingerprint |
UWBMeasure.outlatex(S) |
measurement output latex |
29.8 Utility Functions¶
mesname(n, dirname[, h]) |
get the measurement data filename |
ptw1() |
return W1 Tx and Rx points |
visibility() |
determine visibility type of WHERE1 measurements campaign points |
trait(filename[, itx, dtype, dico, h]) |
evaluate various parameters for all measures in itx array |
29.9 Functions¶
linspace(start, stop[, num, endpoint, …]) |
Return evenly spaced numbers over a specified interval. |
mesname(n, dirname[, h]) |
get the measurement data filename |
polyfit(x, y, deg[, rcond, full, w, cov]) |
Least squares polynomial fit. |
polyval(p, x) |
Evaluate a polynomial at specific values. |
ptw1() |
return W1 Tx and Rx points |
trait(filename[, itx, dtype, dico, h]) |
evaluate various parameters for all measures in itx array |
visibility() |
determine visibility type of WHERE1 measurements campaign points |
29.10 Classes¶
CAL_DATA(d) |
ch1 ch2 ch3 ch4 vna_att1 vna_att2 vna_freq |
FP(M, k[, alpha, Tint, sym, nint, thlos, …]) |
Fingerprint class |
Fdd(d) |
Frequency Domain Deconv Data |
PyLayers |
Generic PyLayers Meta Class |
RAW_DATA(d) |
Members ——- ch1 ch2 ch3 ch4 time timeTX tx |
TFP() |
Tx Rx distance los_cond |
Tdd(d) |
Time Domain Deconv Data |
UWBMeasure([nTx, h, display]) |
UWBMeasure class |
29.11 Class Inheritance Diagram¶
30 pylayers.measures.mesmimo Module¶
30.1 Classes¶
ADPchannel([x, dtype, y, dtype, az, dtype, …]) |
Angular Delay Profile channel |
AFPchannel([x, dtype, y, dtype, tx, dtype, …]) |
Angular Frequency Profile channel |
AntArray(**kwargs) |
Class AntArray |
Array(p[, w]) |
Array class |
Axes3D(fig[, rect]) |
3D axes object. |
Bsignal([x, dtype, y, dtype, label]) |
Signal with an embedded time base |
BuiltinFunctionType |
alias of builtins.builtin_function_or_method |
BuiltinMethodType |
alias of builtins.builtin_function_or_method |
CodeType |
alias of builtins.code |
Combiner(Wbr, Whb, Wsh) |
|
CoroutineType |
alias of builtins.coroutine |
Ctilde() |
container for the 4 components of the polarimetric ray channel |
DynamicClassAttribute([fget, fset, fdel, doc]) |
Route attribute access on a class to __getattr__. |
Error |
|
FBsignal([x, dtype, y, dtype, label]) |
FBsignal : Base signal in Frequency domain |
FHsignal([x, dtype, y, dtype, label]) |
FHsignal : Hermitian uniform signal in Frequency domain |
FUsignal([x, dtype, y, dtype, label]) |
FUsignal : Uniform signal in Frequency Domain |
FrameType |
alias of builtins.frame |
FunctionType |
alias of builtins.function |
GeneratorType |
alias of builtins.generator |
GetSetDescriptorType |
alias of builtins.getset_descriptor |
LambdaType |
alias of builtins.function |
Layout([arg]) |
Handling Layout |
MIMO(**kwargs) |
This class handles the data coming from a MIMO Channel Sounder |
MappingProxyType |
alias of builtins.mappingproxy |
Mchannel(x, y, **kwargs) |
Handle the measured channel |
MemberDescriptorType |
alias of builtins.member_descriptor |
Mesh5([_filename]) |
Class handling hdf5 measurement files |
MethodType |
alias of builtins.method |
ModuleType |
alias of builtins.module |
Noise([ti, tf, fsGHz, PSDdBmpHz, NF, R, seed]) |
Create noise |
PolyCollection(verts[, sizes, closed]) |
|
Precoder(Fhs, Fbh, Fht) |
|
PyLayers |
Generic PyLayers Meta Class |
SCPI([port, timeout, verbose, Nr, Nt, emulated]) |
|
SimpleNamespace |
A simple attribute-based namespace. |
TBchannel([x, dtype, y, dtype, label]) |
radio channel in non uniform delay domain |
TBsignal([x, dtype, y, dtype, label]) |
Based signal in Time domain |
TUDchannel([x, dtype, y, dtype, taud, …]) |
Uniform channel in Time domain with delay |
TUchannel([x, dtype, y, dtype, label]) |
Uniform channel in delay domain |
TUsignal([x, dtype, y, dtype, label]) |
Uniform signal in time domain |
TXRU() |
Tranceiver Units |
Tchannel([x, y, tau, shape, dtype, dod, …]) |
Handle the transmission channel |
TracebackType |
alias of builtins.traceback |
UCArray(p[, w]) |
Uniform Circular Array |
ULArray(**kwargs) |
Uniform Linear Array |
Usignal([x, dtype, y, dtype, label]) |
Signal with an embedded uniform Base |
VLayout |
30.2 Class Inheritance Diagram¶

31 pylayers.measures.cormoran Module¶
This module handles CORMORAN measurement data
31.1 CorSer Class¶
-
class
pylayers.measures.cormoran.CorSer(serie=6, day=11, source='CITI', layout=False)[source]¶ Handle CORMORAN measurement data
Hikob data handling from CORMORAN measurement campaign
- 11/06/2014
- single subject (Bernard and Nicolas)
- 12/06/2014
- several subject (Jihad, Eric , Nicolas)
-
align(devdf, hkbdf)[source]¶ DEPRECATED align time of 2 data frames:
the time delta of the second data frame is applyied on the first one (e.g. time for devdf donwsampled by hkb data frame time)
devdf : device dataframe hkbdf : hkbdataframe
- devdfc :
- aligned copy device dataframe
- hkbdfc :
- aligned copy hkbdataframe
>>> from pylayers.measures.cormoran import * >>> S=CorSer(6) >>> devdf = S.devdf[S.devdf['id']=='HKB:15'] >>> hkbdf = S.hkb['AP1-AnkleLeft'] >>> devdf2,hkbdf2 = S.align(devdf,hkbdf)
-
animhkb(a, b, interval=10, save=False)[source]¶ a : node name |number b : node name | number save : bool
-
animhkbAP(a, AP_list, interval=1, save=False, **kwargs)[source]¶ a : node name AP_nb=[] save : bool
>>> from pylayers.measures.cormoran import * >>> S = CorSer(6) >>> S.animhkbAP('TorsoTopLeft',['AP1','AP2','AP3','AP4'],interval=100,xstart=58,figsize=(20,2))
-
ant¶ display device techno, id , id on body, body owner,…
-
compute_visibility(techno='HKB', square_mda=True, all_links=True)[source]¶ determine visibility of links for a given techno
- techno string
- select the given radio technology of the nodes to determine
- the visibility matrix
- square_mda boolean
- select ouput format
- True : (device x device x timestamp) False : (link x timestamp)
- all_links : bool
- compute all links or just those for which data is available
if square_mda = True
- intersection : (ndevice x nbdevice x nb_timestamp)
- matrice of intersection (1 if link is cut 0 otherwise)
- links : (nbdevice)
- name of the links
if square_mda = False
- intersection : (nblink x nb_timestamp)
- matrice of intersection (1 if link is cut 0 otherwise)
- links : (nblink x2)
- name of the links
>>> from pylayers.measures.cormoran import * >>> import matplotlib.pyplot as plt >>> C=CorSer(serie=14,day=12) >>> inter,links=C.compute_visibility(techno='TCR',square_mda=True) >>> inter.shape (15, 15, 12473) >>>C.imshowvisibility_i(inter,links)
-
dev¶ display device techno, id , id on body, body owner,…
-
devmapper(a, techno='')[source]¶ - retrieve name of device if input is number
- or retrieve number of device if input is name
- a : string
- dev name
- ia : int
- dev number
- ba : string
- dev refernce in body
- subject : string
- body owning the device
-
export_csv(**kwargs)[source]¶ export to csv devices positions
- unit : string (‘mm’|’cm’|’m’),
- unit of positions in csv(default mm)
- tunit: string
- time unit in csv (default ‘ns’)
- ‘alias’: dict
- dictionnary to replace name of the devices into the csv . example : if you want to replace a device id named ‘TCR:34’ to an id = 5, you have to add an entry in the alias dictionnary as : alias.update({‘TCR34’:5})
- offset : np.array
- apply an offset on positions
a csv file into the folder <PylayersProject>/netsave
-
getdevp(a, techno='', t='', fId='')[source]¶ get a device position
- a : str | int
- name |id
- techno : str
- radio techno
optional :
- t : float | list
- given time |[time_start,time_stop]
pa : np.array()
>>> from pylayers.measures.cormoran import * >>> S=CorSer(serie=34) >>> a=S.getdevp('AP1','WristLeft')
-
getlink(a, b, techno='', t='')[source]¶ get a link value
optional :
- techno : str
- radio techno
- t : float | list
- given time or [start,stop] time
Pandas Serie
>>> from pylayers.measures.cormoran import * >>> S=CorSer(serie=34) >>> S.getlink('AP1','WristLeft')
-
getlinkd(a, b, techno='', t='')[source]¶ get the distance for a link between devices
optional
- techno : str
- radio techno
- t : float | list
- given time or [start,stop] time
- dist : np.array()
- all distances for all timestamps for the given link
>>> from pylayers.measures.cormoran import * >>> S = CorSer(serie=6) >>> d = S.getlinkd('AP1','WristLeft',techno='HKB')
-
getlinkp(a, b, technoa='', technob='', t='', fId='')[source]¶ get a link devices positions
optional :
- technoa : str
- radio techno
- technob : str
- radio techno
- t : float | list
- given time | [time_start,time_stop]
OR
- fId : int
- frame id
pa,pb : np.array()
>>> from pylayers.measures.cormoran import * >>> S=CorSer(serie=34) >>> a,b=S.getlinkp('AP1','WristLeft')
-
imshowvisibility(techno='HKB', t=0, **kwargs)[source]¶ imshow visibility mda
techno : (HKB|TCR) t : float
time in second>>> from pylayers.measures.cormoran import * >>> import matplotlib.pyplot as plt >>> C=CorSer(serie=6,day=12) >>> inter,links=C.compute_visibility(techno='TCR',square_mda=True) >>> i,l=C.imshowvisibility_i(inter,links)
pylayers.measures.CorSer.compute_visibility()
-
imshowvisibility_i(techno='HKB', t=0, **kwargs)[source]¶ imshow visibility mda interactive
inter : (nb link x nb link x timestamps) links : (nblinks) time : intial time (s)
>>> from pylayers.measures.cormoran import * >>> import matplotlib.pyplot as plt >>> C=CorSer(serie=6,day=12) >>> inter,links=C.visimda(techno='TCR',square_mda=True) >>> i,l=C.imshowvisibility_i(inter,links)
-
lk2nd(lk)[source]¶ transcode a lk from Id to real name
lk : string
>>> C=Corser(6) >>> lk = 'HKB:15-HKB:7' >>> C.lk2nd(lk)
-
mtlbsave()[source]¶ Matlab format save
- S{day}_{serie}
node_name node_place node_coord
HKB.{linkname}.tr HKB.{linkname}.rssi HKB.{linkname}.td HKB.{linkname}.dist HKB.{linkname}.sh HKB.{linkname}.dsh
TCR.{linkname}.tr HKB.{linkname}.range HKB.{linkname}.td HKB.{linkname}.dist HKB.{linkname}.sh
-
plot(a, b, techno='', t='', **kwargs)[source]¶ ploting
- a : str | int
- name |id
- b : str | int
- name |id
- techno : str (optional)
- radio techno
- t : float | list (optional)
- given time or [start,stop] time
color : color distance : boolean (False)
plot distance instead of value- lin : boolean (False)
- display linear value instead of dB
- sqrtinv : boolean (False)
- apply : “sqrt (1/ dataset)”
- xoffset : float (0)
- add an offset on x axis
- yoffset : float (1|1e3|1e6)
- add an offset on y axis
- title : boolean (True)
- display title
- shortlabel : boolean (True)
- enable short labelling
- fontsize : int (18)
- font size
- returnlines : boolean
- if True return the matplotlib ploted lines
>>> from pylayers.measures.cormoran import * >>> S = CorSer(6) >>> f,ax = S.plot('AP1','TorsoTopLeft',techno='HKB') >>> f,ax = S.pltvisi('AP1','TorsoTopLeft',techno='HKB',fig=f,ax=ax) >>> #f,ax = S.pltmob(fig=f,ax=ax) >>> #plt.title('hatch = visibility / gray= mobility') >>> plt.show()
-
pltgt(a, b, **kwargs)[source]¶ plt ground truth
t0 t1 fig ax figsize: tuple linestyle’ inverse :False,
display 1/distance instead of distance- log : boolean
- display log for distance intead of distance
- gammma’:1.,
- mulitplication factor for log : gamma*log(distance) this can be used to fit RSS
- mode : string
- ‘HKB’ | ‘TCR’ | ‘FULL’
- visi : boolean,
- display visibility
- color: string color (‘k’|’m’|’g’),
- color to display the visibility area
- hatch’: strin hatch type (‘//’)
- hatch type to hatch visibility area
- fontsize: int
- title fontsize
>>> from pylayers.measures.cormoran import * >>> S=CorSer(6) >>> S.pltgt('AP1','TorsoTopLeft')
-
plthkb(a, b, techno='HKB', **kwargs)[source]¶ plot Hikob devices
DEPRECATED
a : node name |number b : node name | number t0 : start time t1 : stop time
>>> from pylayers.measures.cormoran import * >>> S = CorSer(6) >>> f,ax = S.plthkb('AP1','TorsoTopLeft',techno='HKB') >>> f,ax = S.pltvisi('AP1','TorsoTopLeft',techno='HKB',fig=f,ax=ax) >>> f,ax = S.pltmob(fig=f,ax=ax) >>> plt.title('hatch = visibility / gray= mobility') >>> plt.show()
-
pltlk(a, b, **kwargs)[source]¶ plot links
- a : string
- node a name
- b : string
- node b name
- display: list
- techno to be displayed
figsize t0: float
time start- t1 : float
- time stop
colhk: plt.color
color of hk curve- colhk2:plt.color
- color of hk curve2 ( if recirpocal)
- linestylehk:
- linestyle hk
- coltcr:
- color tcr curve
- coltcr2:
- color of tcr curve2 ( if recirpocal)
- linestyletcr:
- linestyle tcr
- colgt:
- color ground truth
- inversegt:
- invert ground truth
- loggt: bool
- apply a log10 factor to ground truth
- gammagt:
- applly a gamma factor to ground truth (if loggt ! )
- fontsize:
- font size of legend
- visi:
- display visibility indicator
- axs :
- list of matplotlib axes
>>> from pylayers.measures.cormoran import * >>> S=CorSer(6) >>> S.pltlk('AP1','TorsoTopLeft')
-
pltmob(**kwargs)[source]¶ plot mobility
- subject: str
- subject to display () if ‘’, take the fist one from self.subject)
- showvel : boolean
- display filtered velocity
- velth: float (0.7)
- velocity threshold
- fo : int (5)
- filter order
- fw: float (0.02)
- 0 < fw < 1 (fN <=> 1)
- time_offset : int
- add time_offset to start later
>>> from pylayers.measures.cormoran import * >>> S = CorSer(6) >>> f,ax = S.plthkb('AP1','TorsoTopLeft',techno='HKB') >>> #f,ax = S.pltvisi('AP1','TorsoTopLeft',techno='HKB',fig=f,ax=ax) >>> f,ax = S.pltmob(fig=f,ax=ax) >>> plt.title('hatch = visibility / gray= mobility') >>> plt.show()
-
plttcr(a, b, **kwargs)[source]¶ plot TCR devices
a : node name |number b : node name | number t0 : start time t1 : stop time
-
pltvisi(a, b, techno='', **kwargs)[source]¶ plot visibility between link a and b
- color:
- fill color
- hatch:
- hatch type
- label_pos: (‘top’|’bottom’|’‘)
- postion of the label
- label_pos_off: float
- offset of postion of the label
- label_mob: str
- prefix of label in mobility
- label_stat: str
- prefix of label static
>>> from pylayers.measures.cormoran import * >>> S = CorSer(6) >>> f,ax = S.plthkb('AP1','TorsoTopLeft',techno='HKB') >>> f,ax = S.pltvisi('AP1','TorsoTopLeft',techno='HKB',fig=f,ax=ax) >>> f,ax = S.pltmob(fig=f,ax=ax) >>> plt.title('hatch = visibility / gray= mobility') >>> plt.show()
-
showlink(a='AP1', b='BackCenter', technoa='HKB', technob='HKB', **kwargs)[source]¶ show link configuation for a given frame
- a : int
- link index
- b : int
- link index
- technoa : string
- default ‘HKB’|’TCR’|’BS’
- technob
- default ‘HKB’|’TCR’|’BS’
- phi : float
- antenna elevation in rad
-
showpattern(a, techno='HKB', **kwargs)[source]¶ show pattern configuation for a given link and frame
- a : int
- link index
- technoa : string
- ‘HKB’|’TCR’|’BS’
- technob
- default ‘HKB’|’TCR’|’BS’
- phi : float
- antenna elevation in rad
fig : ax : t : float phi : float
pi/2ap : boolean
-
snapshot(t0=0, offset=15.5, title=True, save=False, fig=[], ax=[], figsize=(10, 10))[source]¶ single snapshot plot
t0: float offset : float title : boolean save : boolean fig ax figsize : tuple
-
visidev(a, b, technoa='HKB', technob='HKB', dsf=10)[source]¶ get link visibility status
- visi : pandas Series
- 0 : LOS 1 : NLOS
31.1.1 Notes¶
Useful members
distdf : distance between radio nodes (122 columns) devdf : device data frame
31.2 Functions¶
ChangeBasis(u0, v0, w0, v1) |
change basis |
ExpFunc(x, y) |
exponential fitting Parameters ———- x : np.array y : np.array |
Global_Trajectory(cycle, traj) |
global trajectory |
InvFunc(x, z) |
inverse fitting |
LegFunc(nn, ntrunc, theta, phi) |
Compute Legendre functions Ylm(theta,phi) |
PolygonPatch(polygon, **kwargs) |
Constructs a matplotlib patch from a geometric object |
PowFunc(x, y) |
power fitting |
array(object[, dtype, copy, order, subok, ndmin]) |
Create an array. |
cascaded_union |
Returns the union of a sequence of geometries |
cdf(x[, color, label, lw, xlabel, ylabel, logx]) |
plot the cumulative density function of x |
coldict() |
Color dictionary html color |
compint(linterval, zmin, zmax[, tol]) |
get complementary intervals |
cor_log([short]) |
display cormoran measurement campaign logfile |
corrcy(a, b) |
cyclic matching correlation |
cpu_count |
Returns the number of CPUs in the system |
createtrxfile(_filename, freq, phi, theta, …) |
Create antenna trx file Usage:createtrxfile(filename,freq,phi,theta,Fpr,Fpi,Ftr,Fti) |
cshift(l, offset) |
ndarray circular shift Parameters ———- l : ndarray offset : int |
delay(p1, p2) |
calculate delay in ns between 2 points |
dimcmp(ar1, ar2) |
compare shape of arrays |
dist(A, B) |
evaluate the distance between two points A and B |
dist_sh2rssi(dist, Ssh[, offsetdB]) |
Parameters ———- |
encodmtlb(lin) |
encode python list of string in Matlab format |
extract_block_diag(A, M[, k]) |
Extracts blocks of size M from the kth diagonal of square matrix A, whose size must be a multiple of M. |
fill_block_diag(A, blocks, M[, k]) |
fill A with blocks of size M from the kth diagonal |
fill_block_diagMDA(A, blocks, M[, k]) |
fill A with blocks of size M from the kth diagonal |
foo(var1, var2[, long_var_name]) |
A one-line summary that does not use variable names or the function name. |
getdir(longname) |
get directory of a long name |
getlong(shortname, directory) |
get a long name |
getshort(longname) |
get a short name |
img_as_ubyte(image[, force_copy]) |
Convert an image to 8-bit unsigned integer format. |
in_ipynb() |
check if program is run in ipython notebook |
ininter(ar, val1, val2) |
in interval |
lt2idic(lt) |
convert list of tuple to dictionary |
make_axes_locatable(axes) |
|
nbint(a) |
calculate the number of distinct contiguous sets in a sequence of integer |
npextract(y, s) |
access a numpy MDA while keeping number of axis |
outputGi_func(args) |
|
outputGi_func_test(args) |
|
pbar(verbose, **kwargs) |
|
printout(text[, colour]) |
|
randcol(Nc) |
get random color |
read_gpickle(path) |
Read graph object in Python pickle format. |
rgb(valex[, out]) |
convert a hexadecimal color into a (r,g,b) array >>> import pylayers.util.pyutil as pyu >>> coldic = pyu.coldict() >>> val = rgb(coldic[‘gold’],’float’) |
rotate_line(A, B, theta) |
rotation of a line [AB] of an angle theta with A fixed |
rotation(cycle[, alpha]) |
rotate a cycle of frames by an angle alpha |
sqrte(z) |
Evanescent SQRT for waves problems |
time2npa(lt) |
convert pd.datetime.time to numpy array |
timestamp(now) |
|
translate(cycle, new_origin) |
rotate a cycle of frames by an angle alpha |
tstincl(ar1, ar2) |
test wheteher ar1 interval is included in interval ar2 |
untie(a, b) |
Parameters ———- a : np.array b : np.array |
unzipd(path, zipfilename) |
unzip a zipfile to a folder |
unzipf(path, filepath, zipfilename) |
unzip a file from zipfile to a folder |
urlopen(url[, data, timeout, cafile, …]) |
|
writeDetails(t[, description, location]) |
write MeasurementsDetails.txt |
write_gpickle(G, path[, protocol]) |
Write graph in Python pickle format. |
writemeca(ID, time, p, v, a) |
write mecanic information into text file: output/TruePosition.txt output/UWBSensorMeasurements.txt |
writenet(net, t) |
write network information into text file: netsave/ZIGLinkMeasurements.txt netsave/UWBLinkMeasurements.txt |
writenode(agent) |
write Nodes.txt |
zipd(path, zipfilename) |
add a folder to a zipfile |
31.3 Classes¶
Axes3D(fig[, rect]) |
3D axes object. |
Basemap([llcrnrlon, llcrnrlat, urcrnrlon, …]) |
|
Body([_filebody, _filemocap, _filewear, …]) |
Class to manage a Body model |
Button(ax, label[, image, color, hovercolor]) |
A GUI neutral button. |
CheckButtons(ax, labels, actives) |
A GUI neutral radio button. |
CorSer([serie, day, source, layout]) |
Handle CORMORAN measurement data |
Cursor(ax[, horizOn, vertOn, useblit]) |
A horizontal and vertical line that spans the axes and moves with the pointer. |
DF([b, a]) |
Digital Filter Class |
Layout([arg]) |
Handling Layout |
Pool |
alias of pathos.multiprocessing.ProcessPool |
PyLayers |
Generic PyLayers Meta Class |
SelectL(L, fig, ax) |
Associates a Layout and a figure |
Slider(ax, label, valmin, valmax[, valinit, …]) |
A slider representing a floating point range. |
VTKDataSource |
This source manages a VTK dataset given to it. |
combinations |
combinations(iterable, r) –> combinations object |
partial |
partial(func, *args, **keywords) - new function with partial application of the given arguments and keywords. |
product |
product(*iterables, repeat=1) –> product object |
31.4 Class Inheritance Diagram¶

32 pylayers.measures.vna.E5072A Module¶
32.1 Classes¶
BuiltinFunctionType |
alias of builtins.builtin_function_or_method |
BuiltinMethodType |
alias of builtins.builtin_function_or_method |
CodeType |
alias of builtins.code |
CoroutineType |
alias of builtins.coroutine |
DynamicClassAttribute([fget, fset, fdel, doc]) |
Route attribute access on a class to __getattr__. |
FrameType |
alias of builtins.frame |
FunctionType |
alias of builtins.function |
GeneratorType |
alias of builtins.generator |
GetSetDescriptorType |
alias of builtins.getset_descriptor |
LambdaType |
alias of builtins.function |
MappingProxyType |
alias of builtins.mappingproxy |
MemberDescriptorType |
alias of builtins.member_descriptor |
Mesh5([_filename]) |
Class handling hdf5 measurement files |
MethodType |
alias of builtins.method |
ModuleType |
alias of builtins.module |
PyLayers |
Generic PyLayers Meta Class |
SCPI([port, timeout, verbose, Nr, Nt, emulated]) |
|
SimpleNamespace |
A simple attribute-based namespace. |
TracebackType |
alias of builtins.traceback |
32.2 Class Inheritance Diagram¶

33 pylayers.measures.parker.smparker Module¶
33.1 Functions¶
array(object[, dtype, copy, order, subok, ndmin]) |
Create an array. |
bench |
Run benchmarks for module using nose. |
coroutine(func) |
Convert regular generator function to a coroutine. |
fft(a[, n, axis, norm]) |
Compute the one-dimensional discrete Fourier Transform. |
fft2(a[, s, axes, norm]) |
Compute the 2-dimensional discrete Fourier Transform |
fftfreq(n[, d]) |
Return the Discrete Fourier Transform sample frequencies. |
fftn(a[, s, axes, norm]) |
Compute the N-dimensional discrete Fourier Transform. |
fftshift(x[, axes]) |
Shift the zero-frequency component to the center of the spectrum. |
fmin(func, x0[, args, xtol, ftol, maxiter, …]) |
Minimize a function using the downhill simplex algorithm. |
gettty() |
get tty and handles port conflicts |
hfft(a[, n, axis, norm]) |
Compute the FFT of a signal which has Hermitian symmetry (real spectrum). |
ifft(a[, n, axis, norm]) |
Compute the one-dimensional inverse discrete Fourier Transform. |
ifft2(a[, s, axes, norm]) |
Compute the 2-dimensional inverse discrete Fourier Transform. |
ifftn(a[, s, axes, norm]) |
Compute the N-dimensional inverse discrete Fourier Transform. |
ifftshift(x[, axes]) |
The inverse of fftshift. |
ihfft(a[, n, axis, norm]) |
Compute the inverse FFT of a signal which has Hermitian symmetry. |
irfft(a[, n, axis, norm]) |
Compute the inverse of the n-point DFT for real input. |
irfft2(a[, s, axes, norm]) |
Compute the 2-dimensional inverse FFT of a real array. |
irfftn(a[, s, axes, norm]) |
Compute the inverse of the N-dimensional FFT of real input. |
k2xyz(ik, sh) |
Parameters ———- |
loadmat(file_name[, mdict, appendmat]) |
Load MATLAB file. |
make_axes_locatable(axes) |
|
new_class(name[, bases, kwds, exec_body]) |
Create a class object dynamically using the appropriate metaclass. |
prepare_class(name[, bases, kwds]) |
Call the __prepare__ method of the appropriate metaclass. |
rfft(a[, n, axis, norm]) |
Compute the one-dimensional discrete Fourier Transform for real input. |
rfft2(a[, s, axes, norm]) |
Compute the 2-dimensional FFT of a real array. |
rfftfreq(n[, d]) |
Return the Discrete Fourier Transform sample frequencies (for usage with rfft, irfft). |
rfftn(a[, s, axes, norm]) |
Compute the N-dimensional discrete Fourier Transform for real input. |
sleep(seconds) |
Delay execution for a given number of seconds. |
svd(a[, full_matrices, compute_uv]) |
Singular Value Decomposition. |
test |
Run tests for module using nose. |
weights(nx, nz, kx, kz, Kx, Kz) |
Practical Demonstration of Limited Feedback Beamforming for mmWave Systems |
xyztok(iz, iy, ix[, Nx, Ny]) |
33.2 Classes¶
ADPchannel([x, dtype, y, dtype, az, dtype, …]) |
Angular Delay Profile channel |
AFPchannel([x, dtype, y, dtype, tx, dtype, …]) |
Angular Frequency Profile channel |
AntArray(**kwargs) |
Class AntArray |
Array(p[, w]) |
Array class |
Axes([_id, name, ser, scale, typ]) |
This class allows the control of axes |
Axes3D(fig[, rect]) |
3D axes object. |
BuiltinFunctionType |
alias of builtins.builtin_function_or_method |
BuiltinMethodType |
alias of builtins.builtin_function_or_method |
CodeType |
alias of builtins.code |
Combiner(Wbr, Whb, Wsh) |
|
CoroutineType |
alias of builtins.coroutine |
Ctilde() |
container for the 4 components of the polarimetric ray channel |
DynamicClassAttribute([fget, fset, fdel, doc]) |
Route attribute access on a class to __getattr__. |
FrameType |
alias of builtins.frame |
FunctionType |
alias of builtins.function |
GeneratorType |
alias of builtins.generator |
GetSetDescriptorType |
alias of builtins.getset_descriptor |
LambdaType |
alias of builtins.function |
MappingProxyType |
alias of builtins.mappingproxy |
Mchannel(x, y, **kwargs) |
Handle the measured channel |
MemberDescriptorType |
alias of builtins.member_descriptor |
Mesh5([_filename]) |
Class handling hdf5 measurement files |
MethodType |
alias of builtins.method |
ModuleType |
alias of builtins.module |
Precoder(Fhs, Fbh, Fht) |
|
Profile(**kwargs) |
|
PyLayers |
Generic PyLayers Meta Class |
SCPI([port, timeout, verbose, Nr, Nt, emulated]) |
|
Scanner([port, anchors, reset, vel, acc]) |
This class handles the FACS (Four Axes Channel Scanner) |
Serial([port, baudrate, bytesize, parity, …]) |
Serial port class POSIX implementation. |
SimpleNamespace |
A simple attribute-based namespace. |
TBchannel([x, dtype, y, dtype, label]) |
radio channel in non uniform delay domain |
TUDchannel([x, dtype, y, dtype, taud, …]) |
Uniform channel in Time domain with delay |
TUchannel([x, dtype, y, dtype, label]) |
Uniform channel in delay domain |
TXRU() |
Tranceiver Units |
Tchannel([x, y, tau, shape, dtype, dod, …]) |
Handle the transmission channel |
TracebackType |
alias of builtins.traceback |
UCArray(p[, w]) |
Uniform Circular Array |
ULArray(**kwargs) |
Uniform Linear Array |
33.3 Class Inheritance Diagram¶
34 pylayers.signal.bsignal Module¶
34.1 Bsignal Class¶
-
class
pylayers.signal.bsignal.Bsignal(x=array([], dtype=float64), y=array([], dtype=float64), label=[])[source]¶ Signal with an embedded time base
This class gathers a 1D signal and its axis indexation.
The x base is not necessarily uniform
x has 1 axis
x and the last axes of y have the same length
By construction shape(y)[-1] :=len(x), len(x) takes priority in case of observed conflict
-
cformat(**kwargs)[source]¶ complex format
- sax : list
- selected output axis from y default [0,1] typ : string ‘m’ : modulus ‘v’ : value ‘l10’ : dB (10 log10) ‘l20’ : dB (20 log10) ‘d’ : phase degrees ‘r’ : phase radians ‘du’ : phase degrees unwrap ‘ru’ : phase radians unwrap ‘gdn’ : group delay (ns) ‘gdm’ : group distance (m) ‘re’ : real part ‘im’ : imaginary part
- sel : list of ndarray()
- data selection along selected axis, all the axis void default [[],[]]
ik : fixed axis value default (0)
- a0 : first data axis
- this axis can be self.x or not
a1 : second data axis dt : data
- ylabels : string
- label for the selected complex data format
This function returns 2 arrays x and y and the corresponding labels Convention : the last axis of y has same dimension as x y can have an arbitrary number of axis i.e a MIMO channel matrix could be x : f and y : t x r x f
>>> import numpy as np >>> S = Bsignal() >>> x = np.arange(100) >>> y = np.arange(400).reshape(2,2,100)+1j*np.arange(400).reshape(2,2,100) >>> S.x = x >>> S.y = y >>> S.cformat() (array([[-240. , 43.01029996], [ 49.03089987, 52.55272505]]), 'Magnitude (dB)')
-
extract(u)[source]¶ extract a subset of signal from index
u : np.array
O : Usignal
>>> from pylayers.signal.bsignal import * >>> import numpy as np >>> x = np.arange(0,1,0.01) >>> y = np.sin(2*np.pi*x) >>> s = Bsignal(x,y) >>> su = s.extract(np.arange(4,20)) >>> f,a = s.plot() >>> f,a = su.plot()
-
flatteny(yrange=[], reversible=False)[source]¶ flatten y array
yrange : array of y index values to be flattenned reversible : boolean
if True the sum is place in object member yf else y is smashed
-
gating(xmin, xmax)[source]¶ gating between xmin and xmax
xmin : float xmax : float
nothing self is modified>>> import numpy as np >>> import matplotlib.pyplot as plt >>> from pylayers.signal.bsignal import * >>> x = np.linspace(-10,10,100) >>> y = np.sin(2*np.pi*12*x)+np.random.normal(0,0.1,len(x)) >>> s = TUsignal(x,y) >>> fig,ax = s.plot(typ=['v']) >>> txt1 = plt.title('before gating') >>> plt.show()(Source code, png, hires.png, pdf)
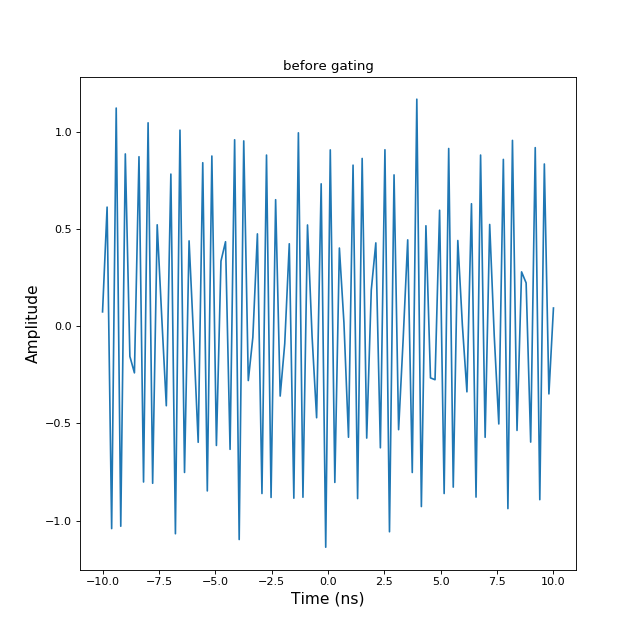
>>> s.gating(-3,4) >>> fig,ax=s.plot(typ=['v']) >>> txt2 = plt.title('after gating') >>> plt.show()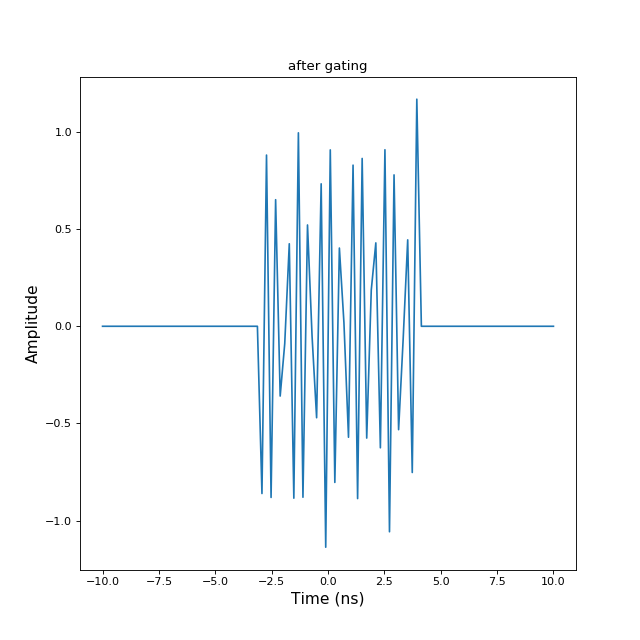 When a gating is applied the removed information is lost
When a gating is applied the removed information is lost
-
imshow(**kwargs)[source]¶ imshow of y matrix
- interpolation : string
- ‘none’|’nearest’|’bilinear’
- cmap : colormap
- plt.cm.jet
- aspect : string
- ‘auto’ (default) ,’equal’,’scalar’
- function : string
- {‘imshow’|’pcolormesh’}
- typ : string
- ‘l20’,’l10’
vmin : min value vmax : max value sax : list
select axebindex
>>> f = np.arange(100) >>> y = np.random.randn(50,100)+1j*np.random.randn(50,100) >>> F = FUsignal(f,y)
-
plot(**kwargs)[source]¶ plot signal Bsignal
iy : index of the waveform to plot (-1 = all) col : string
default ‘black’vline : ndarray hline : ndarray unit1 : string
default ‘V’- unit2 : string
- default ‘V’
xmin : float xmax : float ax : axes instance or [] dB : boolean
default False- dist : boolean
- default False
- display : boolean
- default True
- logx : boolean
- defaut False
- logy : boolean
- default False
fig : figure ax : np.array of axes
pylayers.util.plotutil.mulcplot
-
save(filename)[source]¶ save Bsignal in Matlab File Format
filename : string
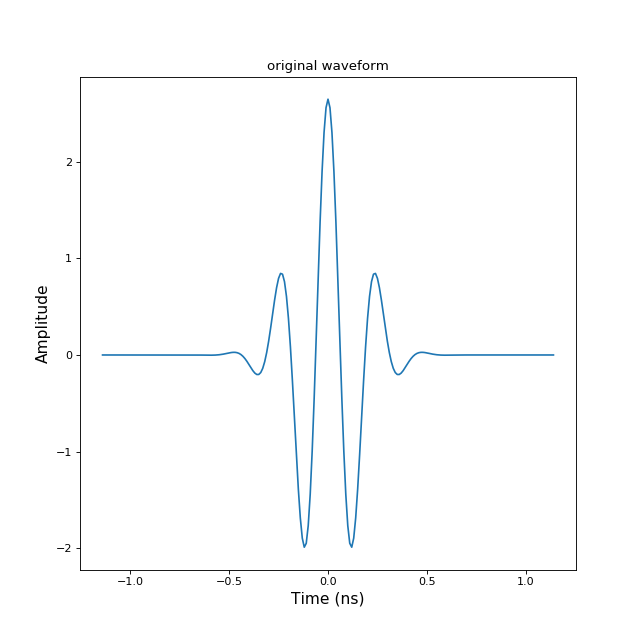
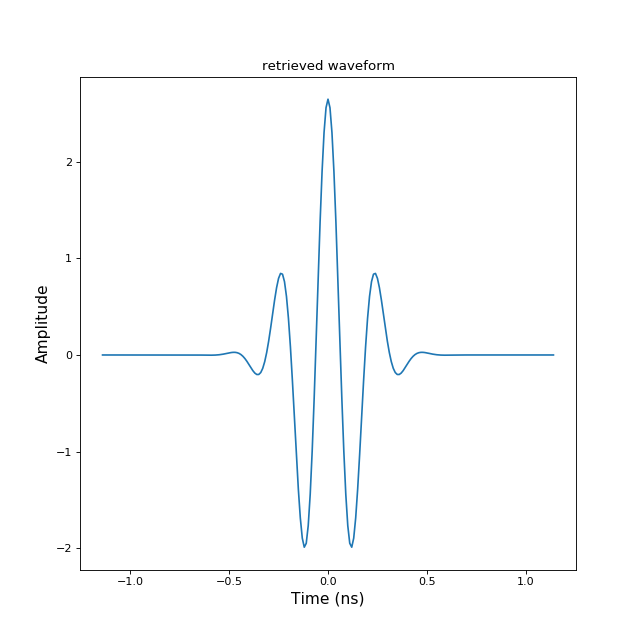
>>> from pylayers.signal.bsignal import * >>> import matplotlib.pyplot as plt >>> e = TUsignal() >>> e.EnImpulse(feGHz=100) >>> fig,ax = e.plot(typ=['v']) >>> tit1 = plt.title('original waveform') >>> e.save('impulse.mat') >>> del e >>> h = TUsignal() >>> h.load('impulse.mat') >>> fig,ax = h.plot(typ=['v']) >>> tit2 = plt.title('retrieved waveform')Bsignal.load
-
34.2 Usignal Class¶
34.3 TBsignal Class¶
-
class
pylayers.signal.bsignal.TBsignal(x=array([], dtype=float64), y=array([], dtype=float64), label=[])[source]¶ Based signal in Time domain
-
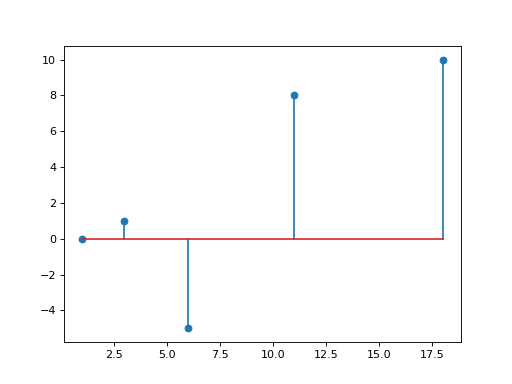
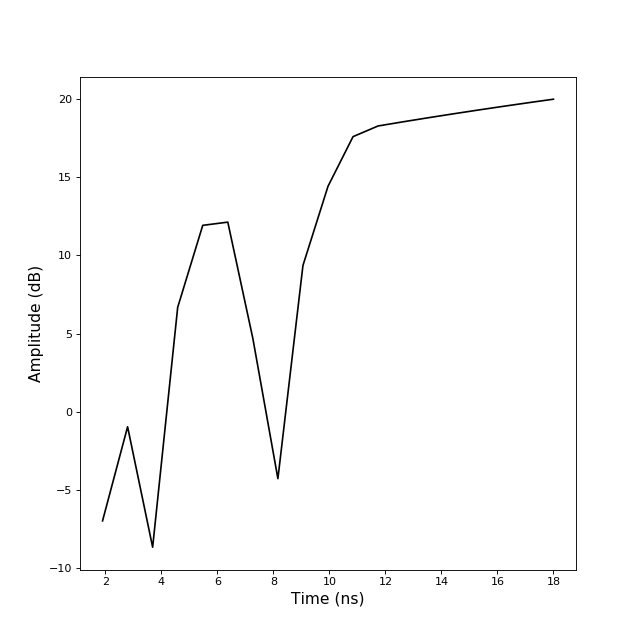
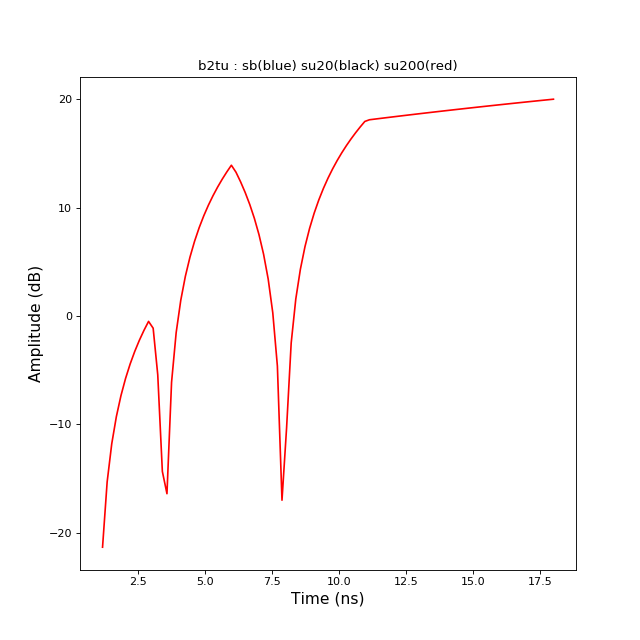
b2tu(N)[source]¶ conversion from TBsignal to TUsignal
- N : integer
- Number of points
U : TUsignal
This function exploits linear interp1d
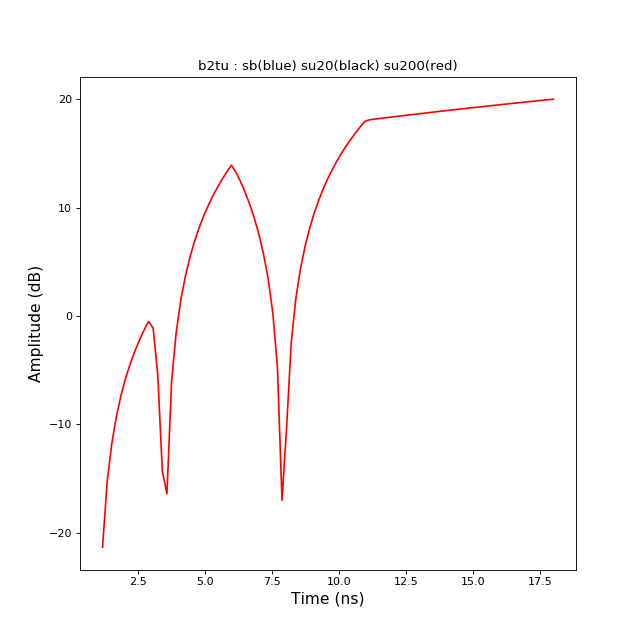
>>> from pylayers.signal.bsignal import * >>> import matplotlib.pyplot as plt >>> x = np.array( [ 1, 3 , 6 , 11 , 18]) >>> y = np.array( [ 0,1 ,-5, 8 , 10]) >>> sb = TBsignal(x,y) >>> su20 = sb.b2tu(20) >>> su100 = sb.b2tu(100) >>> fi = plt.figure() >>> st = sb.stem() >>> fig,ax = su20.plot(color='k') >>> fig,ax = su100.plot(color='r') >>> ti = plt.title('b2tu : sb(blue) su20(black) su200(red)')
-
b2tu2(fsGHz, Tobsns)[source]¶ conversion from TBsignal to TUsignal
- N : integer
- Number of points
U : TUsignal
This function exploits linear interp1d
>>> from pylayers.signal.bsignal import * >>> import matplotlib.pyplot as plt >>> x = np.array( [ 1, 3 , 6 , 11 , 18]) >>> y = np.array( [ 0,1 ,-5, 8 , 10]) >>> sb = TBsignal(x,y) >>> su20 = sb.b2tu(20) >>> su100 = sb.b2tu(100) >>> fi = plt.figure() >>> st = sb.stem() >>> fig,ax = su20.plot(color='k') >>> fig,ax = su100.plot(color='r') >>> ti = plt.title('b2tu : sb(blue) su20(black) su200(red)')
-
integ(Tns, Tsns=50)[source]¶ integation of alphak tauk of TBsignal
used energy detector for IEEE 802.15.6 standard
Tns : Tsns :
-
plot(**kwargs)[source]¶ plot TBsignal
unit1 : actual unit of data unit2 : unit for display dist : boolean xmin : float xmax : float logx : boolean logy :boolean
>>> import numpy as np >>> from pylayers.signal.bsignal import * >>> from matplotlib.pylab import * >>> x = np.array( [ 1, 3 , 6 , 11 , 18]) >>> y = np.array( [ 0,1 ,-5, 8 , 10]) >>> s = Bsignal(x,y) >>> fi = figure() >>> fig,ax=s.plot(typ=['v']) >>> ti = title('TBsignal : plot') >>> show()
-
tap(fcGHz, WGHz, Ntap)[source]¶ Back to baseband
- fcGHz : float
- center frequency
- WGHz : float
- bandwidth
- Ntap : int
- Number of tap
Implement formula (2.52) from D.Tse book page 50
-
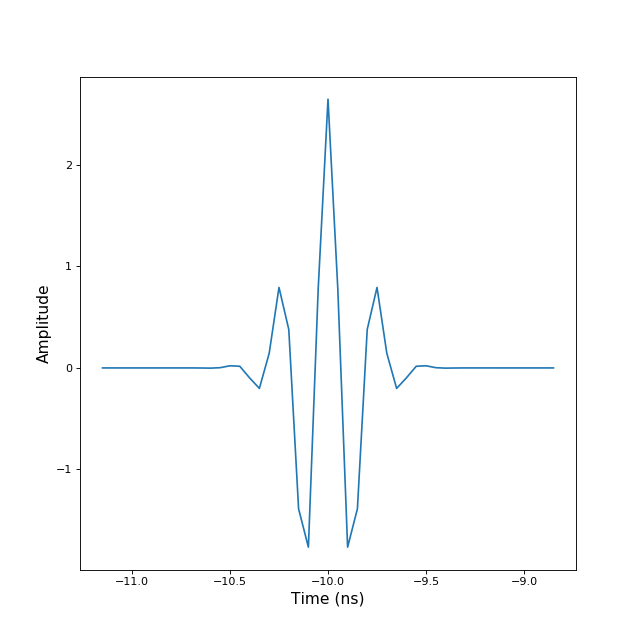
translate(tau)[source]¶ translate TBsignal signal by tau
- tau : float
- delay for translation
Once translated original signal and translated signal might not be on the same grid
>>> from pylayers.signal.bsignal import * >>> from matplotlib.pylab import * >>> ip = TUsignal() >>> ip.EnImpulse() >>> ip.translate(-10) >>> fig,ax=ip.plot(typ=['v']) >>> show()
(Source code, png, hires.png, pdf)
-
34.4 TUsignal Class¶
-
class
pylayers.signal.bsignal.TUsignal(x=array([], dtype=float64), y=array([], dtype=float64), label=[])[source]¶ Uniform signal in time domain
This class inheritates from TBsignal and Usignal
The signal can either be complex or real
-
EnImpulse(**kwargs)[source]¶ Create an energy normalized Gaussian impulse (Usignal)
fcGHz : float WGHz : float threshdB : float feGHz : float
-
Impulse(**kwargs)[source]¶ Create a Gaussian impulse (Usignal)
fcGHz : float WGHz : float threshdB : float feGHz : float
-
Yadd_zeros2l(N)[source]¶ time domain extension on the left with N zeros
- N : integer
- number of additinal zero values
Yadd_zeros2r
Work only for single y
-
Yadd_zeros2r(N)[source]¶ time domain extension on right with N zeros
- N : integer
- number of additinal zero values
Yadd_zeros2l
-
align(u2)[source]¶ align two TUsignal on a same base
returns a list which contains the two aligned signals
It is assume that both signal u1 and u2 share the same dx This function can be improved regarding time granularity
u2 : TUsignal
TUsignal y extended TU signal, x bases are adjusted
>>> import matplotlib.pylab as plt >>> from pylayers.signal.bsignal import * >>> i1 = TUsignal() >>> i2 = TUsignal() >>> i1.EnImpulse() >>> i2.EnImpulse() >>> i2.translate(-10) >>> i3 = i1.align(i2) >>> fig,ax=i3.plot(typ=['v']) >>> plt.show()
-
correlate(s, normalized=True)[source]¶ correlates with an other TUsignal
s : TUsignal normalized : boolean
default TrueThe time step dx should be the same
The chosen time reference is the one of the self signal This means that this function can be used to detect events appearing in the self signal by correlating with signal ..math:s, maximum of the correlation function appears in the proper time of the self signal history.
Interpretation of the correlation levels
We assume that s1 is the self signal and s2 is the correlating signal
s1 = a1 * sn(t-tau) where sn is normalized in energy s2 = a2 * sn(t) ” ” ” ” “
the maximum of correlation appears at time tau and its value is
max (s1.correlate(s2,True)) = a1*a2- In the case of the propagation simulation we have
- s1 : the time domain UWB received waveform s2 : the corresponding emitted waveform s2,
To determine the ratio a1/a2 or a2/a1 in order to evaluate the propagation Loss L = 20 log10(a2/a1)
This calculus assumes implicitely that the shape of the transmitted signal has not been modified during the propagation, which is a strong hypothesis. This calculus is then a minorant of the received energy.
L = max(s1.correlate(s2,True)/s1.Energy) with a1**2 = sum(s1**s1)*dx
-
corrgauss(sigma)[source]¶ Correlate TUsignal with a normalized gaussian of standard deviation sigma
- sigma : float
- standard deviation of Gaussian
-
esd(mode='bilateral')[source]¶ Calculate the energy spectral density of the U signal
- mode : string
- ‘unilateral’ | ‘bilateral’
FHsignal : if mode == ‘bilateral’ FUsignal : if mode == ‘unilateral’
FHsignal.unrex
-
fft(shift=False)[source]¶ forward fast Fourier transform of TUsignal
- shift : boolean
- default False
This fft is a scaled fft and takes into account the value of the sampling period.
FHsignal
>>> from pylayers.signal.bsignal import * >>> e = TUsignal() >>> e.EnImpulse() >>> E = e.fft()
-
fftsh()[source]¶ returns an FHsignal
This fft is a scaled fft and takes into account the value of the sampling period.
FHsignal : Frequency signal with enforced Hermitian symetry
-
filter(order=4, wp=0.3, ws=0.4, ftype='butter')[source]¶ TUsignal filtering
order : int wp : float ws : float ftype : string
O : Output filtered TUsignal
-
ftshift()[source]¶ return the associated FUsignal
H : FUsignal
pylayers.signal.bsignal.TUsignal.fftsh pylayers.signal.waveform.ip_generic
-
psd(Tpns=100, R=50, periodic=True)[source]¶ calculate TUsignal power spectral density
R : Resistance (default 50 Ohms) Tpns : real
PRP (default 100 ns)Note
If time is in ns the resulting PSD is expressed in dBm/MHz (~10-9)
-
34.5 FBsignal Class¶
-
class
pylayers.signal.bsignal.FBsignal(x=array([], dtype=float64), y=array([], dtype=float64), label=[])[source]¶ FBsignal : Base signal in Frequency domain
plot : plot modulus and phase plotri : plot real part and imaginary part plotdB : plot modulus in dB
-
plot(**kwargs)[source]¶ plot FBsignal
- phase : boolean
- default True
- dB : boolean
- default True
iy : index of y value to be displayed typ : string
[‘l10’,’m,’l20’,’d’,’r’,’du’,’ru’]xlabels ylabels
>>> from pylayers.signal.bsignal import * >>> from numpy import * >>> from scipy import * >>> S = FBsignal() >>> S.x = arange(100) >>> S.y = cos(2*pi*S.x)+1j*sin(3*pi*S.x+pi/3) >>> fig,ax = S.plot() >>> plt.show()
Bsignal.plot pylayers.util.plotutil.mulcplot
-
34.6 FUsignal Class¶
-
class
pylayers.signal.bsignal.FUsignal(x=array([], dtype=float64), y=array([], dtype=float64), label=[])[source]¶ FUsignal : Uniform signal in Frequency Domain
x : nd.array((1xN)) y : Complex nd.array((… x N )
symH : force Hermitian symetry –> FHsignal symHz : force Hermitian symetry with zero padding –> FHsignal align : align two FUsignal on a same frequency base enthrsh : Energy thresholding thresh = 99.99 % dBthrsh : dB thresholding thresh = 40dB ift : Inverse Fourier transform resample : resampling with a new base newdf : resampling with a new df zp : zero padding until len(x) = N plotri : plot real part and imaginary part plotdB : plot modulus in dB get : get k th ray window :
-
applyFriis()[source]¶ apply Friis factor
The Friis factor is multiplied to y
\[ \begin{align}\begin{aligned}y := \( \frac{-j c}{4 \pi f} \) y\\x is frequency in GHz\end{aligned}\end{align} \]boolean isFriis is set to True
-
energy(axis=-1, Friis=False, mode='mean')[source]¶ calculate energy along a given axis of the FUsignal
axis : (default 0) Friis : boolean mode : string
mean (default) | center | integ | first | last>>> ip = TUsignal() >>> ip.EnImpulse() >>> En1 = ip.energy() >>> assert((En1>0.99) & (En1<1.01))
If Friis is true energy is multiplied by
\(\frac{-j*c}{4 pi fGHz}\)
axis = 0 is ray axis
if mode == ‘mean’
\(E=\frac{1}{K} \sum_k |y_k|^2\)
if mode == ‘integ’
\(E=\delta_x \sum_k |y_k|^2\)
if mode == ‘center’
\(E= |y_{K/2}|^2\)
-
enthrsh(thresh=99.99)[source]¶ Energy thresholding of an FUsignal
- thresh : float
- threshold in percentage (default 99.99)
EMH2 : cumul energie H
-
ifft(Npt=-1)[source]¶ Inverse Fourier transform
- Npt : int
- default -1 (same number as x)
tc : TUsignal
\[x = \textrm{linspace}(0,\frac{1}{\delta f},N)\]\[y = [0,\mathbb{X}_u^T]\]
-
ift(Nz=1, ffts=0, beta=0)[source]¶ Inverse Fourier Transform - returns the associated TUsignal
Nz : Number of zeros (-1) No forcing ffts : 0 (no fftshift 1:fftshift) beta : kaiser window shape
0 : rectangular 5 : similar to a Hamming 6 : similar to a Hanning 8.6 : similar to a blackmanuh :
>>> from pylayers.signal.bsignal import * >>> e = TUsignal() >>> e.EnImpulse() >>> E = e.fft() >>> EU = E.unrex()
1 - Force Hermitian symmetry –> FHsignal with or without zero padding 2 - Inverse Fourier transform with or without fftshift
FHsignal.ifft, FUsignal.symH, FUsignal.symHz
-
iftshift(Nz=1)[source]¶ Return the associated TUsignal
apply the inverse fftshift operator to come back in time
-
symHz(Nz, scale='extract')[source]¶ Force Hermitian symmetry with zero padding
- Nz : int
- Number of zero above f[-1]
- scale : string
- default : ‘extract’ | ‘cir’
SH : FHsignal
The signal is rescaled in order to conserve energy
Let denotes the FUsignal as \(mathcal{X}_d\) The Fourier matrix is :math:`left[ matrix{mathbb{1}\
mathbb{W}_d\ mathbb{W}_u}right]`FHSignal
>>> N = 11 >>> x = np.linspace(2,11,N) >>> y1 = sp.randn(N)+1j*sp.rand(N) >>> y2 = sp.randn(N)+1j*sp.rand(N) >>> y1[0] = 0 >>> y2[0] = 0 >>> y = np.vstack((y1,y2)) >>> S = FUsignal(x,y) >>> FH = S.symHz(10)
-
tap(**kwargs)[source]¶ calculates channel taps
- fcGHz : float
- center frequency GHz
- WGHz : float
- bandwidth GHz
Ntap : number of taps baseband : boolean
default : Truehtapi
[Tse] David Tse, http://www.eecs.berkeley.edu/~dtse/Chapters_PDF/Fundamentals_Wireless_Communication_chapter2.pdf page 26
-
34.7 FHsignal Class¶
34.8 Noise Class¶
34.9 Functions¶
ExpFunc(x, y) |
exponential fitting Parameters ———- x : np.array y : np.array |
InvFunc(x, z) |
inverse fitting |
LegFunc(nn, ntrunc, theta, phi) |
Compute Legendre functions Ylm(theta,phi) |
PowFunc(x, y) |
power fitting |
cdf(x[, color, label, lw, xlabel, ylabel, logx]) |
plot the cumulative density function of x |
cformat(x, y, **kwargs) |
complex format |
coldict() |
Color dictionary html color |
compint(linterval, zmin, zmax[, tol]) |
get complementary intervals |
copy(x) |
Shallow copy operation on arbitrary Python objects. |
corrcy(a, b) |
cyclic matching correlation |
correlate(in1, in2[, mode, method]) |
Cross-correlate two N-dimensional arrays. |
createtrxfile(_filename, freq, phi, theta, …) |
Create antenna trx file Usage:createtrxfile(filename,freq,phi,theta,Fpr,Fpi,Ftr,Fti) |
cshift(l, offset) |
ndarray circular shift Parameters ———- l : ndarray offset : int |
cspline1d(signal[, lamb]) |
Compute cubic spline coefficients for rank-1 array. |
cspline1d_eval(cj, newx[, dx, x0]) |
Evaluate a spline at the new set of points. |
cylinder(fig, pa, pb, R) |
plot a cylinder |
deepcopy(x[, memo, _nil]) |
Deep copy operation on arbitrary Python objects. |
delay(p1, p2) |
calculate delay in ns between 2 points |
dimcmp(ar1, ar2) |
compare shape of arrays |
displot(pt, ph[, arrow]) |
discontinuous plot |
encodmtlb(lin) |
encode python list of string in Matlab format |
extract_block_diag(A, M[, k]) |
Extracts blocks of size M from the kth diagonal of square matrix A, whose size must be a multiple of M. |
fill_block_diag(A, blocks, M[, k]) |
fill A with blocks of size M from the kth diagonal |
fill_block_diagMDA(A, blocks, M[, k]) |
fill A with blocks of size M from the kth diagonal |
firwin(numtaps, cutoff[, width, window, …]) |
FIR filter design using the window method. |
foo(var1, var2[, long_var_name]) |
A one-line summary that does not use variable names or the function name. |
getdir(longname) |
get directory of a long name |
getlong(shortname, directory) |
get a long name |
getshort(longname) |
get a short name |
iirdesign(wp, ws, gpass, gstop[, analog, …]) |
Complete IIR digital and analog filter design. |
iirfilter(N, Wn[, rp, rs, btype, analog, …]) |
IIR digital and analog filter design given order and critical points. |
in_ipynb() |
check if program is run in ipython notebook |
ininter(ar, val1, val2) |
in interval |
lfilter(b, a, x[, axis, zi]) |
Filter data along one-dimension with an IIR or FIR filter. |
lt2idic(lt) |
convert list of tuple to dictionary |
mulcplot(x, y, **kwargs) |
handling multiple complex variable plots |
nbint(a) |
calculate the number of distinct contiguous sets in a sequence of integer |
npextract(y, s) |
access a numpy MDA while keeping number of axis |
pol3D(fig, rho, theta, phi[, sf, shade, title]) |
polar 3D surface plot |
polycol(lpoly[, var]) |
plot a collection of polygon |
printout(text[, colour]) |
|
randcol(Nc) |
get random color |
rectplot(x, xpos[, ylim]) |
plot rectangles on an axis |
rgb(valex[, out]) |
convert a hexadecimal color into a (r,g,b) array >>> import pylayers.util.pyutil as pyu >>> coldic = pyu.coldict() >>> val = rgb(coldic[‘gold’],’float’) |
rotate_line(A, B, theta) |
rotation of a line [AB] of an angle theta with A fixed |
shadow(data, ax) |
data : np.array 0 or 1 ax : matplotlib.axes |
shp(arr) |
return dimension of an array |
sqrte(z) |
Evanescent SQRT for waves problems |
test() |
|
timestamp(now) |
|
tstincl(ar1, ar2) |
test wheteher ar1 interval is included in interval ar2 |
untie(a, b) |
Parameters ———- a : np.array b : np.array |
unzipd(path, zipfilename) |
unzip a zipfile to a folder |
unzipf(path, filepath, zipfilename) |
unzip a file from zipfile to a folder |
writeDetails(t[, description, location]) |
write MeasurementsDetails.txt |
writemeca(ID, time, p, v, a) |
write mecanic information into text file: output/TruePosition.txt output/UWBSensorMeasurements.txt |
writenet(net, t) |
write network information into text file: netsave/ZIGLinkMeasurements.txt netsave/UWBLinkMeasurements.txt |
writenode(agent) |
write Nodes.txt |
zipd(path, zipfilename) |
add a folder to a zipfile |
34.10 Classes¶
Axes3D(fig[, rect]) |
3D axes object. |
Bsignal([x, dtype, y, dtype, label]) |
Signal with an embedded time base |
Error |
|
FBsignal([x, dtype, y, dtype, label]) |
FBsignal : Base signal in Frequency domain |
FHsignal([x, dtype, y, dtype, label]) |
FHsignal : Hermitian uniform signal in Frequency domain |
FUsignal([x, dtype, y, dtype, label]) |
FUsignal : Uniform signal in Frequency Domain |
Noise([ti, tf, fsGHz, PSDdBmpHz, NF, R, seed]) |
Create noise |
PolyCollection(verts[, sizes, closed]) |
|
PyLayers |
Generic PyLayers Meta Class |
TBsignal([x, dtype, y, dtype, label]) |
Based signal in Time domain |
TUsignal([x, dtype, y, dtype, label]) |
Uniform signal in time domain |
Usignal([x, dtype, y, dtype, label]) |
Signal with an embedded uniform Base |
34.11 Class Inheritance Diagram¶
35 pylayers.signal.standard Module¶
35.1 Classes¶
AP(**kwargs) |
Access Point |
Band(**kwargs) |
A Band is a structured portion of the spectrum |
Channel(fcghz, bmhz[, gmhz]) |
a radio channel abstraction |
PyLayers |
Generic PyLayers Meta Class |
Wstandard([stdname, _filejson]) |
Wireless standard class |
35.2 Class Inheritance Diagram¶
36 pylayers.signal.device Module¶
This module describes the radio devices to be used for electromagentic simulations
36.1 Device Class¶
Device.__init__([devname, ID, owner, typ]) |
init of Device class |
36.3 Classes¶
Antenna([typ]) |
Attributes ———- |
Device([devname, ID, owner, typ]) |
Device Class |
PyLayers |
Generic PyLayers Meta Class |
VTKDataSource |
This source manages a VTK dataset given to it. |
Wstandard([stdname, _filejson]) |
Wireless standard class |
36.4 Class Inheritance Diagram¶
37 pylayers.signal.DF Module¶
37.1 DF Class¶
This class implements a LTI digital filter
-
class
pylayers.signal.DF.DF(b=array([1]), a=array([1]))[source]¶ Digital Filter Class
- a : array
- transfer function coefficients denominator
- b : array
- transfer function coefficients numerator
- p : array dtype=complex
- transfer function poles
- z : array dtype=complex
- transfer function zeros
filter : filter a signal freqz : Display transfer function ellip_bp : elliptic bandpath filter remez : FIR design
-
butter(order=5, w=0.25, typ='low')[source]¶ Butterwoth digital filter design
- order : int
- filter order
- w : array_like
- a scalar or length-2 sequence (relative frequency 0<1 fN <=> 1)
- typ : string
- ‘lowpass’ | ‘highpass’ | ‘bandpass’ | ‘bandstop’
>>> from pylayers.signal.DF import * >>> flt = DF() >>> flt.butter(order=3,w=0.25,typ='low') >>> flt.freqz()
-
ellip_bp(wp, ws, gpass=0.5, gstop=20)[source]¶ Elliptic Bandpath filter
wp : ws : gpass gstop :
iirdesign
-
factorize()[source]¶ factorize the filter into a minimal phase and maximal phase
- (Fmin,Fmax) : tuple of filter
- Fmin : minimal phase filter Fmax : maximal phase filter
-
filter(s, causal=True)[source]¶ filter s
- s : np.array |TUsignal
- input signal
- y : np.array | TUsignal
- output signal
>>> import matplotlib.pyplot as plt >>> df = DF(b=np.array([1,1],a=np.array([1,-1])) >>> N = 100 >>> s = np.zeros(N) >>> s[0] = 1 >>> y = df.filter(s) >>> plt.stem(np.arange(N),x) >>> plt.stem(np.arange(N),y) >>> plt.show()
-
freqz(**kwargs)[source]¶ freqz : evaluation of filter transfer function
The function evaluates an FUsignal
- display : boolean
- True
- fsGHz : float
- if set to 0 (relative frequency)
-
match()[source]¶ return a match filter
DF :
if $$H(z)=\frac{1+b_1z^1+…+b_Nz^{-N}}{1+a_1z^1+…+a_Mz^{-M}}$$
it returns
$$H_m(z)=\frac{b_N+b_{N-1}z^1+…+z^{-N}}{1+a_1z^1+…+a_Mz^{-M}}$$
-
remez(numtaps=401, bands=(0, 0.1, 0.11, 0.2, 0.21, 0.5), desired=(0.0001, 1, 0.0001))[source]¶ FIR design Remez algorithm
numtaps : int bands : tuple of N floats desired : tuple of N/2 floats
flt.remez(numtaps=401,bands=(0,0.1,0.11,0.2,0.21,0.5),desired=(0.0001,1,0.0001)):
numtaps = 401 bands = (0,0.1,0.11,0.2,0.21,0.5) desired = (0.0001,1,0.0001))
flt.remez( numtaps , bands , desired)
37.3 Class Inheritance Diagram¶

38 pylayers.signal.waveform Module¶
38.2 Class Inheritance Diagram¶

39 pylayers.mobility.agent Module¶
39.1 Agent Class¶
39.2 Classes¶
Agent(**args) |
Class Agent |
Containment |
Class Containment |
Gcom(net, sim) |
Communication graph |
InterpenetrationConstraint |
Class InterpenetrationConstaint |
Layout([arg]) |
Handling Layout |
Localization(**args) |
Handle localization engine of agents |
Network([owner, EMS, PN]) |
Network class |
Node(**kwargs) |
Class Node |
PLocalization([loc, tx, loc_updt_time, sim]) |
|
Person([ID, interval, roomId, L, net, wld, …]) |
Person Process |
RX(**args) |
RX process |
Seek |
class Seek |
Separation |
Class Separation |
TX(**args) |
TX process ( not used for now) Each agent use the TX process for querying LDP/information/message passing data to other agent |
vec3(*args) |
Three-dimensional vector. |
39.3 Class Inheritance Diagram¶

40 pylayers.mobility.ban.body Module¶
40.1 Body Class¶
This class implements the body model
-
class
pylayers.mobility.ban.body.Body(_filebody='John.ini', _filemocap=[], _filewear=[], traj=[], unit='cm', loop=True, centered=True, multi_subject_mocap=False, color='white')[source]¶ Class to manage a Body model
ncyl : number of cylinder sl : d : topos : vtopos :
load center posvel loadC3D settopos setccs setdcs geomfile plot3d movie cylinder_basis_k cyl_antenna
-
anim()[source]¶ animate body
>>> from pylayers.mobility.trajectory import * >>> from pylayers.mobility.ban.body import * >>> from pylayers.gis.layout import * >>> T=Trajectories() >>> T.loadh5() >>> L=Layout(T.Lfilename) >>> B = Body() >>> B.settopos(T[0],t=0,cs=True) >>> L._show3() >>> B.anim(B)
-
animc3d()[source]¶ animate c3d file
>>> from pylayers.mobility.trajectory import * >>> from pylayers.mobility.ban.body import * >>> B = Body() >>> B.animc3d(B)
-
body_link(topos=True, frameId=0)[source]¶ body link
- topos : boolean
- default True
- frameId : int
- used in case topos == False. Indicates the frame Id.
- link_vis : np.array (,nlinks)
- number of intersected cylinder on the link
links is a list of couple of strings inticating the different links between devices.
-
center(force=False)[source]¶ centering the body
self.pg : center of gravity self.vg : velocity self.d : set of centered frames self.smocap : integrated distance self.vmocap : averaged velocity
The center method creates a centered version of the motion capture data stored in self.d It also calculates : self.smocap : total distance along trajectory self.vmocap : averaged speed along trajectory
Here only the projection of the body centroid in the plan 0xy is calculated
-
chronophoto(**kwargs)[source]¶ - chronophotography of the body movement
- (a.k.a. position as a function of time)
- tstart : float
- starting time in second
- tend : float
- ending time in second,
- tstep : float
- time step between tstart and tend
- sstep : float
- spatial step (distance between 2 instant)
- planes : list
- list of planes to be displayed [‘xz’,’xy’,’yz’]
- dev : bool
- show devices
- dev : bool
- show devices ids
-
cyl_antenna(cylinderId, l, alpha, frameId=0)[source]¶ cylinder antenna
cylinderId : int index of cylinder l : distance from origin of cylider alpha : angle from reference direction frameId : frameId
-
dpdf(tr=[], tunit='ns', poffset=False)[source]¶ device position dataframe return a dataframe with body and devices positions along the self.traj
- tr : ndarray
- timerange
- cdf: pd.DataFrame
- complete device data frame
>>> from pylayers.mobility.ban.body import * >>> T = tr.Trajectories() >>> T.loadh5() >>> B=Body(traj=T[0]) >>> cdf = B.dpdf()
-
export_csv(unit='mm', df=[], _filename='default.csv', col=['dev_id', 'dev_x', 'dev_y', 'dev_z', 'timestamp'], **kwargs)[source]¶
-
geomfile(**kwargs)[source]¶ create a geomview file from a body configuration
iframe : int frame id (useless if topos==True) verbose : boolean topos : boolean frame id or topos wire : boolean body as a wire or cylinder ccs : boolean display cylinder coordinate system cacs : boolean display cylinder antenna coordinate system acs : boolean display antenna coordinate system struc : boolean displat structure layout tag : string filestruc : string name of the Layout
This function creates either a 3d representation of the frame iframe or if topos==True a representation of the current topos.
-
getdevT(id=-1, t=[], frameId=[])[source]¶ get device orientation
- id : str | list
- device id.
- frameId : int
- frameid
- t : float
- time
device orientation
-
getdevp(id=-1)[source]¶ get device position
- id : str | list
- device id.
- frameId : int
- frameid
- t : float
- time
device position
-
intersectBody(A, B, topos=True, frameId=0, cyl=[])[source]¶ intersect Body
A : np.array (3,) B : np.array (3,) topos : boolean frameId : 0 cyl : list
exclusion list- intersect : np.array (,ncyl)
- O : AB not intersected by cylinder 1 : AB intersected by cylinder
-
intersectBody3(A, B, topos=True, frameId=0)[source]¶ intersect body new version
A B topos frameId cyl
- intersect : np.array (,ncyl)
- O : AB not intersected by cylinder 1 : AB intersected by cylinder
-
load(_filebody='John.ini', _filemocap=[], unit=[], _filewear=[])[source]¶ load a body ini file
_filebody : body short filename
A body .ini file contains 4 sections
- section [nodes]
Node number = Node name + section [cylinder] CylinderId = {‘t’:tail node number, ‘h’:head node number , ‘r’: cylinder’ radius} + section [wearable] + section [mocap]
-
loadC3D(filename='07_01.c3d', nframes=-1, unit='cm')[source]¶ load nframes of motion capture C3D file
- filename : string
- file name
- nframes : int
- number of frames
- unit : str (mm|cm|mm
- unit of c3d file
- rot : list [‘x’,’y’,’z’]
- swap axes of the c3d file
-
movie(**kwargs)[source]¶ creates a geomview movie
lframe : [] verbose : False topos : True wire : True ccs : False dcs : False struc : True traj : [] filestruc:’DLR.off’
Body.geomfile
-
network()[source]¶ evaluate network topology and dynamics
This function evaluates distance, velocity and acceleration of the radio network nodes
self.D2 : distances between radio nodes self.V2 : velocities between radio nodes self.A2 : accelerations between radio nodes
-
plot3d(iframe=0, topos=False, fig=[], ax=[], col='b')[source]¶ scatter 3d plot
iframe : int topos : boolean fig : ax : col : string
fig,ax
-
posvel(traj, t)[source]¶ calculate position and velocity
- traj : Tajectory DataFrame
- nx3
- t : float
- trajectory time for evaluation of topos
kf : frame integer index kt : trajectory integer index vsn : normalized speed vector along motion capture trajectory (source) wsn : planar vector orthogonal to vsn vtn : normalized speed vector along motion trajectory (target) wtn : planar vector orthogonal to wtn
This funtion takes as argument a trajectory which is a panda dataframe and a time value in the time scale of the trajectory.
t value should of course be in the interval between trajecroty extremal times tmin and tmax.
smax is the maximum distance covered in the whole motion capture sequence.
sk is the distance covered from the begining of the trajectory until the trajectory time t.
The ratio between those 2 distance is rounded to the nearest integer.
:math:`delta = s_k -lceil
rac{s_k}{s_{max}} s_{max}`
k_f is the index of the topos motion capture into the MOCAP
- |__________|__________|____________|___________|_____
- kf=2
tmin tmax 0 smax sk
-
setccs(frameId=0, topos=False)[source]¶ set cylinder coordinate system
frameId : int frame id in the mocap dataframe (default 0) topos : boolean default False
self.ccs : ndarray (nc,3,3) Notes —–
There are as many frames as cylinders (body graph edges)
ccs is a MDA (nc x 3 x 3 ) where nc denotes the number of cylinders For each cylinder there is an attached coordinate systems
1st vector 2nd
-
setcs(topos=True, frameId=0)[source]¶ set coordinates systems from a topos or a frame id
- topos : boolean
- default : True
- frameId : int
- default 0
pylayers.mobility.ban.body.setccs() pylayers.mobility.ban.body.setdcs() pylayers.mobility.ban.body.setacs()
-
setdcs(topos=True, frameId=0)[source]¶ set device coordinate system (dcs) from a topos
This method evaluates the set of all dcs. It provides the information necessary for device placement on the body.
If N is the number of antenna an dcs is an MDA of size 3x4xN
- topos : boolean
- default : True
- frameId : int
- default 0
self.dcs : dictionnary
>>> import numpy as np >>> import pylayers.mobility.trajectory as tr >>> import pylayers.mobility.ban.body as body >>> import matplotlib.pyplot as plt >>> time = np.arange(0,10,0.1) >>> v = 4000/3600. >>> x = v*time >>> y = np.zeros(len(time)) >>> traj = tr.Trajectory() >>> traj.generate() >>> bc = body.Body() >>> bc.settopos(traj,2.3,2) >>> bc.setccs(topos=True) >>> bc.setdcs() >>> bc.show(plane='yz',color='b',widthfactor=80) >>> plt.show()
-
settopos(traj=[], t=0, cs=True, treadmill=False, p0=array([ 0., 0.]))[source]¶ translate the body on a time stamped trajectory
traj : ndarray (3,N) t,x,y t : float time for evaluation of topos (seconds) this value should be in the range of the trajectory timestamp
self.topos self.vtopos
>>> import numpy as np >>> import pylayers.mobility.trajectory as tr >>> import pylayers.mobility.ban.body as body >>> import matplotlib.pyplot as plt >>> time = np.arange(0,10,0.1) >>> v = 4000/3600. >>> x = v*time >>> y = np.zeros(len(time)) >>> traj = tr.Trajectory() >>> traj.generate() >>> John = body.Body() >>> John.settopos(traj,2.3) >>> fig,ax = John.show(plane='xz',color='b') >>> plt.title('xz') >>> plt.show()topos is the current spatial global position of a body configuration. this method takes as argument a trajectory and a time value t in the trajectory time-scale.
pylayers.util.geomutil.affine
-
40.2 Cylinder Class¶
40.3 Functions¶
ChangeBasis(u0, v0, w0, v1) |
change basis |
Global_Trajectory(cycle, traj) |
global trajectory |
dist(A, B) |
evaluate the distance between two points A and B |
rotation(cycle[, alpha]) |
rotate a cycle of frames by an angle alpha |
translate(cycle, new_origin) |
rotate a cycle of frames by an angle alpha |
40.4 Classes¶
Axes3D(fig[, rect]) |
3D axes object. |
Body([_filebody, _filemocap, _filewear, …]) |
Class to manage a Body model |
PyLayers |
Generic PyLayers Meta Class |
40.5 Class Inheritance Diagram¶

41 pylayers.measures.cormoran Module¶
This module handles CORMORAN measurement data
41.1 CorSer Class¶
-
class
pylayers.measures.cormoran.CorSer(serie=6, day=11, source='CITI', layout=False)[source] Handle CORMORAN measurement data
Hikob data handling from CORMORAN measurement campaign
- 11/06/2014
- single subject (Bernard and Nicolas)
- 12/06/2014
- several subject (Jihad, Eric , Nicolas)
-
align(devdf, hkbdf)[source] DEPRECATED align time of 2 data frames:
the time delta of the second data frame is applyied on the first one (e.g. time for devdf donwsampled by hkb data frame time)
devdf : device dataframe hkbdf : hkbdataframe
- devdfc :
- aligned copy device dataframe
- hkbdfc :
- aligned copy hkbdataframe
>>> from pylayers.measures.cormoran import * >>> S=CorSer(6) >>> devdf = S.devdf[S.devdf['id']=='HKB:15'] >>> hkbdf = S.hkb['AP1-AnkleLeft'] >>> devdf2,hkbdf2 = S.align(devdf,hkbdf)
-
animhkb(a, b, interval=10, save=False)[source] a : node name |number b : node name | number save : bool
-
animhkbAP(a, AP_list, interval=1, save=False, **kwargs)[source] a : node name AP_nb=[] save : bool
>>> from pylayers.measures.cormoran import * >>> S = CorSer(6) >>> S.animhkbAP('TorsoTopLeft',['AP1','AP2','AP3','AP4'],interval=100,xstart=58,figsize=(20,2))
-
ant display device techno, id , id on body, body owner,…
-
compute_visibility(techno='HKB', square_mda=True, all_links=True)[source] determine visibility of links for a given techno
- techno string
- select the given radio technology of the nodes to determine
- the visibility matrix
- square_mda boolean
- select ouput format
- True : (device x device x timestamp) False : (link x timestamp)
- all_links : bool
- compute all links or just those for which data is available
if square_mda = True
- intersection : (ndevice x nbdevice x nb_timestamp)
- matrice of intersection (1 if link is cut 0 otherwise)
- links : (nbdevice)
- name of the links
if square_mda = False
- intersection : (nblink x nb_timestamp)
- matrice of intersection (1 if link is cut 0 otherwise)
- links : (nblink x2)
- name of the links
>>> from pylayers.measures.cormoran import * >>> import matplotlib.pyplot as plt >>> C=CorSer(serie=14,day=12) >>> inter,links=C.compute_visibility(techno='TCR',square_mda=True) >>> inter.shape (15, 15, 12473) >>>C.imshowvisibility_i(inter,links)
-
dev display device techno, id , id on body, body owner,…
-
devmapper(a, techno='')[source] - retrieve name of device if input is number
- or retrieve number of device if input is name
- a : string
- dev name
- ia : int
- dev number
- ba : string
- dev refernce in body
- subject : string
- body owning the device
-
export_csv(**kwargs)[source] export to csv devices positions
- unit : string (‘mm’|’cm’|’m’),
- unit of positions in csv(default mm)
- tunit: string
- time unit in csv (default ‘ns’)
- ‘alias’: dict
- dictionnary to replace name of the devices into the csv . example : if you want to replace a device id named ‘TCR:34’ to an id = 5, you have to add an entry in the alias dictionnary as : alias.update({‘TCR34’:5})
- offset : np.array
- apply an offset on positions
a csv file into the folder <PylayersProject>/netsave
-
getdevp(a, techno='', t='', fId='')[source] get a device position
- a : str | int
- name |id
- techno : str
- radio techno
optional :
- t : float | list
- given time |[time_start,time_stop]
pa : np.array()
>>> from pylayers.measures.cormoran import * >>> S=CorSer(serie=34) >>> a=S.getdevp('AP1','WristLeft')
-
getlink(a, b, techno='', t='')[source] get a link value
optional :
- techno : str
- radio techno
- t : float | list
- given time or [start,stop] time
Pandas Serie
>>> from pylayers.measures.cormoran import * >>> S=CorSer(serie=34) >>> S.getlink('AP1','WristLeft')
-
getlinkd(a, b, techno='', t='')[source] get the distance for a link between devices
optional
- techno : str
- radio techno
- t : float | list
- given time or [start,stop] time
- dist : np.array()
- all distances for all timestamps for the given link
>>> from pylayers.measures.cormoran import * >>> S = CorSer(serie=6) >>> d = S.getlinkd('AP1','WristLeft',techno='HKB')
-
getlinkp(a, b, technoa='', technob='', t='', fId='')[source] get a link devices positions
optional :
- technoa : str
- radio techno
- technob : str
- radio techno
- t : float | list
- given time | [time_start,time_stop]
OR
- fId : int
- frame id
pa,pb : np.array()
>>> from pylayers.measures.cormoran import * >>> S=CorSer(serie=34) >>> a,b=S.getlinkp('AP1','WristLeft')
-
imshow(time=100, kind='time')[source] DEPRECATED
kind : string
‘mean’,’std’
-
imshowvisibility(techno='HKB', t=0, **kwargs)[source] imshow visibility mda
techno : (HKB|TCR) t : float
time in second>>> from pylayers.measures.cormoran import * >>> import matplotlib.pyplot as plt >>> C=CorSer(serie=6,day=12) >>> inter,links=C.compute_visibility(techno='TCR',square_mda=True) >>> i,l=C.imshowvisibility_i(inter,links)
pylayers.measures.CorSer.compute_visibility()
-
imshowvisibility_i(techno='HKB', t=0, **kwargs)[source] imshow visibility mda interactive
inter : (nb link x nb link x timestamps) links : (nblinks) time : intial time (s)
>>> from pylayers.measures.cormoran import * >>> import matplotlib.pyplot as plt >>> C=CorSer(serie=6,day=12) >>> inter,links=C.visimda(techno='TCR',square_mda=True) >>> i,l=C.imshowvisibility_i(inter,links)
-
lk2nd(lk)[source] transcode a lk from Id to real name
lk : string
>>> C=Corser(6) >>> lk = 'HKB:15-HKB:7' >>> C.lk2nd(lk)
-
loadlog()[source] load in self.log the log of the current serie from MeasurementLog.csv
-
mtlbsave()[source] Matlab format save
- S{day}_{serie}
node_name node_place node_coord
HKB.{linkname}.tr HKB.{linkname}.rssi HKB.{linkname}.td HKB.{linkname}.dist HKB.{linkname}.sh HKB.{linkname}.dsh
TCR.{linkname}.tr HKB.{linkname}.range HKB.{linkname}.td HKB.{linkname}.dist HKB.{linkname}.sh
-
offset_setter(a='HKB:1', b='HKB:12', techno='', **kwargs)[source] offset setter
-
offset_setter_video(a='AP1', b='WristRight', **kwargs)[source] video offset setter
-
plot(a, b, techno='', t='', **kwargs)[source] ploting
- a : str | int
- name |id
- b : str | int
- name |id
- techno : str (optional)
- radio techno
- t : float | list (optional)
- given time or [start,stop] time
color : color distance : boolean (False)
plot distance instead of value- lin : boolean (False)
- display linear value instead of dB
- sqrtinv : boolean (False)
- apply : “sqrt (1/ dataset)”
- xoffset : float (0)
- add an offset on x axis
- yoffset : float (1|1e3|1e6)
- add an offset on y axis
- title : boolean (True)
- display title
- shortlabel : boolean (True)
- enable short labelling
- fontsize : int (18)
- font size
- returnlines : boolean
- if True return the matplotlib ploted lines
>>> from pylayers.measures.cormoran import * >>> S = CorSer(6) >>> f,ax = S.plot('AP1','TorsoTopLeft',techno='HKB') >>> f,ax = S.pltvisi('AP1','TorsoTopLeft',techno='HKB',fig=f,ax=ax) >>> #f,ax = S.pltmob(fig=f,ax=ax) >>> #plt.title('hatch = visibility / gray= mobility') >>> plt.show()
-
pltgt(a, b, **kwargs)[source] plt ground truth
t0 t1 fig ax figsize: tuple linestyle’ inverse :False,
display 1/distance instead of distance- log : boolean
- display log for distance intead of distance
- gammma’:1.,
- mulitplication factor for log : gamma*log(distance) this can be used to fit RSS
- mode : string
- ‘HKB’ | ‘TCR’ | ‘FULL’
- visi : boolean,
- display visibility
- color: string color (‘k’|’m’|’g’),
- color to display the visibility area
- hatch’: strin hatch type (‘//’)
- hatch type to hatch visibility area
- fontsize: int
- title fontsize
>>> from pylayers.measures.cormoran import * >>> S=CorSer(6) >>> S.pltgt('AP1','TorsoTopLeft')
-
plthkb(a, b, techno='HKB', **kwargs)[source] plot Hikob devices
DEPRECATED
a : node name |number b : node name | number t0 : start time t1 : stop time
>>> from pylayers.measures.cormoran import * >>> S = CorSer(6) >>> f,ax = S.plthkb('AP1','TorsoTopLeft',techno='HKB') >>> f,ax = S.pltvisi('AP1','TorsoTopLeft',techno='HKB',fig=f,ax=ax) >>> f,ax = S.pltmob(fig=f,ax=ax) >>> plt.title('hatch = visibility / gray= mobility') >>> plt.show()
-
pltlk(a, b, **kwargs)[source] plot links
- a : string
- node a name
- b : string
- node b name
- display: list
- techno to be displayed
figsize t0: float
time start- t1 : float
- time stop
colhk: plt.color
color of hk curve- colhk2:plt.color
- color of hk curve2 ( if recirpocal)
- linestylehk:
- linestyle hk
- coltcr:
- color tcr curve
- coltcr2:
- color of tcr curve2 ( if recirpocal)
- linestyletcr:
- linestyle tcr
- colgt:
- color ground truth
- inversegt:
- invert ground truth
- loggt: bool
- apply a log10 factor to ground truth
- gammagt:
- applly a gamma factor to ground truth (if loggt ! )
- fontsize:
- font size of legend
- visi:
- display visibility indicator
- axs :
- list of matplotlib axes
>>> from pylayers.measures.cormoran import * >>> S=CorSer(6) >>> S.pltlk('AP1','TorsoTopLeft')
-
pltmob(**kwargs)[source] plot mobility
- subject: str
- subject to display () if ‘’, take the fist one from self.subject)
- showvel : boolean
- display filtered velocity
- velth: float (0.7)
- velocity threshold
- fo : int (5)
- filter order
- fw: float (0.02)
- 0 < fw < 1 (fN <=> 1)
- time_offset : int
- add time_offset to start later
>>> from pylayers.measures.cormoran import * >>> S = CorSer(6) >>> f,ax = S.plthkb('AP1','TorsoTopLeft',techno='HKB') >>> #f,ax = S.pltvisi('AP1','TorsoTopLeft',techno='HKB',fig=f,ax=ax) >>> f,ax = S.pltmob(fig=f,ax=ax) >>> plt.title('hatch = visibility / gray= mobility') >>> plt.show()
-
plttcr(a, b, **kwargs)[source] plot TCR devices
a : node name |number b : node name | number t0 : start time t1 : stop time
-
pltvisi(a, b, techno='', **kwargs)[source] plot visibility between link a and b
- color:
- fill color
- hatch:
- hatch type
- label_pos: (‘top’|’bottom’|’‘)
- postion of the label
- label_pos_off: float
- offset of postion of the label
- label_mob: str
- prefix of label in mobility
- label_stat: str
- prefix of label static
>>> from pylayers.measures.cormoran import * >>> S = CorSer(6) >>> f,ax = S.plthkb('AP1','TorsoTopLeft',techno='HKB') >>> f,ax = S.pltvisi('AP1','TorsoTopLeft',techno='HKB',fig=f,ax=ax) >>> f,ax = S.pltmob(fig=f,ax=ax) >>> plt.title('hatch = visibility / gray= mobility') >>> plt.show()
-
savemat()[source] save in Matlab Format
-
showlink(a='AP1', b='BackCenter', technoa='HKB', technob='HKB', **kwargs)[source] show link configuation for a given frame
- a : int
- link index
- b : int
- link index
- technoa : string
- default ‘HKB’|’TCR’|’BS’
- technob
- default ‘HKB’|’TCR’|’BS’
- phi : float
- antenna elevation in rad
-
showpattern(a, techno='HKB', **kwargs)[source] show pattern configuation for a given link and frame
- a : int
- link index
- technoa : string
- ‘HKB’|’TCR’|’BS’
- technob
- default ‘HKB’|’TCR’|’BS’
- phi : float
- antenna elevation in rad
fig : ax : t : float phi : float
pi/2ap : boolean
-
snapshot(t0=0, offset=15.5, title=True, save=False, fig=[], ax=[], figsize=(10, 10))[source] single snapshot plot
t0: float offset : float title : boolean save : boolean fig ax figsize : tuple
-
snapshots(t0=0, t1=10, offset=15.5)[source] take snapshots
t0 : float t1 : float
-
visidev(a, b, technoa='HKB', technob='HKB', dsf=10)[source] get link visibility status
- visi : pandas Series
- 0 : LOS 1 : NLOS
-
visidev2(a, b, technoa='HKB', technob='HKB', trange=[])[source] get link visibility status
- trange : nd array
- time range
- visi : pandas Series
- 0 : LOS 1 : NLOS
-
vlc()[source] play video of the associated serie
41.1.1 Notes¶
Useful members
distdf : distance between radio nodes (122 columns) devdf : device data frame
41.2 Functions¶
ChangeBasis(u0, v0, w0, v1) |
change basis |
ExpFunc(x, y) |
exponential fitting Parameters ———- x : np.array y : np.array |
Global_Trajectory(cycle, traj) |
global trajectory |
InvFunc(x, z) |
inverse fitting |
LegFunc(nn, ntrunc, theta, phi) |
Compute Legendre functions Ylm(theta,phi) |
PolygonPatch(polygon, **kwargs) |
Constructs a matplotlib patch from a geometric object |
PowFunc(x, y) |
power fitting |
array(object[, dtype, copy, order, subok, ndmin]) |
Create an array. |
cascaded_union |
Returns the union of a sequence of geometries |
cdf(x[, color, label, lw, xlabel, ylabel, logx]) |
plot the cumulative density function of x |
coldict() |
Color dictionary html color |
compint(linterval, zmin, zmax[, tol]) |
get complementary intervals |
cor_log([short]) |
display cormoran measurement campaign logfile |
corrcy(a, b) |
cyclic matching correlation |
cpu_count |
Returns the number of CPUs in the system |
createtrxfile(_filename, freq, phi, theta, …) |
Create antenna trx file Usage:createtrxfile(filename,freq,phi,theta,Fpr,Fpi,Ftr,Fti) |
cshift(l, offset) |
ndarray circular shift Parameters ———- l : ndarray offset : int |
delay(p1, p2) |
calculate delay in ns between 2 points |
dimcmp(ar1, ar2) |
compare shape of arrays |
dist(A, B) |
evaluate the distance between two points A and B |
dist_sh2rssi(dist, Ssh[, offsetdB]) |
Parameters ———- |
encodmtlb(lin) |
encode python list of string in Matlab format |
extract_block_diag(A, M[, k]) |
Extracts blocks of size M from the kth diagonal of square matrix A, whose size must be a multiple of M. |
fill_block_diag(A, blocks, M[, k]) |
fill A with blocks of size M from the kth diagonal |
fill_block_diagMDA(A, blocks, M[, k]) |
fill A with blocks of size M from the kth diagonal |
foo(var1, var2[, long_var_name]) |
A one-line summary that does not use variable names or the function name. |
getdir(longname) |
get directory of a long name |
getlong(shortname, directory) |
get a long name |
getshort(longname) |
get a short name |
img_as_ubyte(image[, force_copy]) |
Convert an image to 8-bit unsigned integer format. |
in_ipynb() |
check if program is run in ipython notebook |
ininter(ar, val1, val2) |
in interval |
lt2idic(lt) |
convert list of tuple to dictionary |
make_axes_locatable(axes) |
|
nbint(a) |
calculate the number of distinct contiguous sets in a sequence of integer |
npextract(y, s) |
access a numpy MDA while keeping number of axis |
outputGi_func(args) |
|
outputGi_func_test(args) |
|
pbar(verbose, **kwargs) |
|
printout(text[, colour]) |
|
randcol(Nc) |
get random color |
read_gpickle(path) |
Read graph object in Python pickle format. |
rgb(valex[, out]) |
convert a hexadecimal color into a (r,g,b) array >>> import pylayers.util.pyutil as pyu >>> coldic = pyu.coldict() >>> val = rgb(coldic[‘gold’],’float’) |
rotate_line(A, B, theta) |
rotation of a line [AB] of an angle theta with A fixed |
rotation(cycle[, alpha]) |
rotate a cycle of frames by an angle alpha |
sqrte(z) |
Evanescent SQRT for waves problems |
time2npa(lt) |
convert pd.datetime.time to numpy array |
timestamp(now) |
|
translate(cycle, new_origin) |
rotate a cycle of frames by an angle alpha |
tstincl(ar1, ar2) |
test wheteher ar1 interval is included in interval ar2 |
untie(a, b) |
Parameters ———- a : np.array b : np.array |
unzipd(path, zipfilename) |
unzip a zipfile to a folder |
unzipf(path, filepath, zipfilename) |
unzip a file from zipfile to a folder |
urlopen(url[, data, timeout, cafile, …]) |
|
writeDetails(t[, description, location]) |
write MeasurementsDetails.txt |
write_gpickle(G, path[, protocol]) |
Write graph in Python pickle format. |
writemeca(ID, time, p, v, a) |
write mecanic information into text file: output/TruePosition.txt output/UWBSensorMeasurements.txt |
writenet(net, t) |
write network information into text file: netsave/ZIGLinkMeasurements.txt netsave/UWBLinkMeasurements.txt |
writenode(agent) |
write Nodes.txt |
zipd(path, zipfilename) |
add a folder to a zipfile |
41.3 Classes¶
Axes3D(fig[, rect]) |
3D axes object. |
Basemap([llcrnrlon, llcrnrlat, urcrnrlon, …]) |
|
Body([_filebody, _filemocap, _filewear, …]) |
Class to manage a Body model |
Button(ax, label[, image, color, hovercolor]) |
A GUI neutral button. |
CheckButtons(ax, labels, actives) |
A GUI neutral radio button. |
CorSer([serie, day, source, layout]) |
Handle CORMORAN measurement data |
Cursor(ax[, horizOn, vertOn, useblit]) |
A horizontal and vertical line that spans the axes and moves with the pointer. |
DF([b, a]) |
Digital Filter Class |
Layout([arg]) |
Handling Layout |
Pool |
alias of pathos.multiprocessing.ProcessPool |
PyLayers |
Generic PyLayers Meta Class |
SelectL(L, fig, ax) |
Associates a Layout and a figure |
Slider(ax, label, valmin, valmax[, valinit, …]) |
A slider representing a floating point range. |
VTKDataSource |
This source manages a VTK dataset given to it. |
combinations |
combinations(iterable, r) –> combinations object |
partial |
partial(func, *args, **keywords) - new function with partial application of the given arguments and keywords. |
product |
product(*iterables, repeat=1) –> product object |
41.4 Class Inheritance Diagram¶